Evaluating and Validating Sunflower Reference Genes for Q-PCR Studies Under High Temperature Condition
- PMID: 37228632
- PMCID: PMC10203189
- DOI: 10.30498/ijb.2023.338375.3357
Evaluating and Validating Sunflower Reference Genes for Q-PCR Studies Under High Temperature Condition
Abstract
Background: Q-PCR is the method of choice for PCR- based transcriptomics and validating microarray-based and RNA-seq results. Proper application of this technology requires proper normalization to correct as much as possible errors propagating during RNA extraction and cDNA synthesis.
Objectives: The investigation was performed to find stable reference genes in sunflower under shifting in ambient temperature.
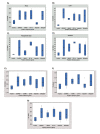
Materials and methods: Sequences of five well-known reference genes of Arabidopsis (Actin, Ubiquitin, Elongation factor-1, GAPDH, and SAND) and one well-known reference gene inhuman, Importin, were subjected to BLASTX against sunflower databases and the relevant genes were subjected to primer designing for q-PCR. Two sunflower inbred lines were cultivated at two dates so that anthesis occurred at nearly 30 °C and 40 °C (heat stress). The experiment was repeated for two years. Q-PCR was run on samples taken for two planting date separately at the beginning of anthesis for each genotype from leaf, taproots, receptacle base, immature and mature disc flowers and on pooled samples comprising of the tissues for each genotype, planting dates and also all tissues for both genotypes and both planting dates. Basic statistical properties of each candidate gene across all the samples were calculated. Furthermore, gene expression stability analysis was done for six candidate reference genes on Cq mean of two years using three independent algorithms, geNorm, Bestkeeper, and Refinder.
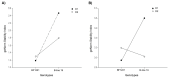
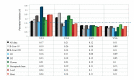
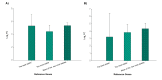
Results: Designed primers for Actin2, SAND, GAPDH, Ubiquitin, EF-1a, and Importin yielded a single peak in melting curve analysis indicating specificity of the PCR reaction. Basic statistical analysis showed that Actin2 and EF-1a had the highest and lowest expression levels across all the samples, respectively. Actin2 appeared to be the most stable reference gene across all the samples based on the three used algorithms. Pairwise variation analysis revealed that for samples taken under ambient temperature of 30 °C, Actin2, EF-1a, SAND and for those taken under ambient temperature of 40 °C, Actin2, EF-1a, Importin and SAND have to be used for normalization in q-PCR studies. Moreover, it is suggested that normalization to be based on Actin2, SAND and EF-1a for vegetative tissues and Actin2, EF-1a, SAND and Importin for reproductive tissues.
Conclusions: In the present research, proper reference genes for normalization of gene expression studies under heat stress conditions were introduced. Moreover, the presence of genotype-by- planting date interaction effects and tissue specific gene expression pattern on the behavior of the most three stable reference genes was indicated.
Keywords: Heat stress; Reference genes; Sunflower.
Copyright: © 2021 The Author(s); Published by Iranian Journal of Biotechnology.
Figures

References
-
- Martins MQ, Fortunato AS, Rodrigues WP, Partelli FL, Campostrini E, Lidon FC, et al. Selection and validation of reference genes for accurate RT-qPCR data normalization in Coffea spp. under a climate changes context of interacting elevated (CO2) and temperature. Front Plant Sci. 2017;8:307. doi: 10.3389/fpls.2017.00307. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
