Combined analysis of metabolome and transcriptome of wheat kernels reveals constitutive defense mechanism against maize weevils
- PMID: 37229118
- PMCID: PMC10204651
- DOI: 10.3389/fpls.2023.1147145
Combined analysis of metabolome and transcriptome of wheat kernels reveals constitutive defense mechanism against maize weevils
Abstract
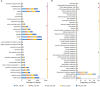
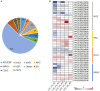
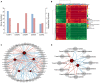
Sitophilus zeamais (maize weevil) is one of the most destructive pests that seriously affects the quantity and quality of wheat (Triticum aestivum L.). However, little is known about the constitutive defense mechanism of wheat kernels against maize weevils. In this study, we obtained a highly resistant variety RIL-116 and a highly susceptible variety after two years of screening. The morphological observations and germination rates of wheat kernels after feeding ad libitum showed that the degree of infection in RIL-116 was far less than that in RIL-72. The combined analysis of metabolome and transcriptome of RIL-116 and RIL-72 wheat kernels revealed differentially accumulated metabolites were mainly enriched in flavonoids biosynthesis-related pathway, followed by glyoxylate and dicarboxylate metabolism, and benzoxazinoid biosynthesis. Several flavonoids metabolites were significantly up-accumulated in resistant variety RIL-116. In addition, the expression of structural genes and transcription factors (TFs) related to flavonoids biosynthesis were up-regulated to varying degrees in RIL-116 than RIL-72. Taken together, these results indicated that the biosynthesis and accumulation of flavonoids contributes the most to wheat kernels defense against maize weevils. This study not only provides insights into the constitutive defense mechanism of wheat kernels against maize weevils, but may also play an important role in the breeding of resistant varieties.
Keywords: Sitophilus zeamais; Triticum aestivum L.; constitutive defense; flavonoids biosynthesis; metabolome; transcriptome.
Copyright © 2023 Lv, Guo, Zhao, Liu, Li and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Efficacy of diatomaceous earth to control internal infestations of rice weevil and maize weevil (Coleoptera: Curculionidae).J Econ Entomol. 2003 Apr;96(2):510-8. doi: 10.1603/0022-0493-96.2.510. J Econ Entomol. 2003. PMID: 14994822
-
Mechanisms of Resistance to Spot Blotch in Yunnan Iron Shell Wheat Based on Metabolome and Transcriptomics.Int J Mol Sci. 2022 May 6;23(9):5184. doi: 10.3390/ijms23095184. Int J Mol Sci. 2022. PMID: 35563578 Free PMC article.
-
Transcriptomic and Metabolomic Analysis of Wheat Kernels in Response to the Feeding of Orange Wheat Blossom Midges (Sitodiplosis mosellana) in the Field.J Agric Food Chem. 2022 Feb 9;70(5):1477-1493. doi: 10.1021/acs.jafc.1c06239. Epub 2022 Jan 28. J Agric Food Chem. 2022. PMID: 35090120
-
Male-produced aggregation pheromone of the maize weevil,Sitophilus zeamais, and interspecific attraction between threeSitophilus species.J Chem Ecol. 1983 Jul;9(7):831-41. doi: 10.1007/BF00987808. J Chem Ecol. 1983. PMID: 24407756
-
Integrated transcriptome and metabolome analysis reveals that flavonoids function in wheat resistance to powdery mildew.Front Plant Sci. 2023 Feb 1;14:1125194. doi: 10.3389/fpls.2023.1125194. eCollection 2023. Front Plant Sci. 2023. PMID: 36818890 Free PMC article.
Cited by
-
Aphid Resistance Evaluation and Constitutive Resistance Analysis of Eighteen Lilies.Insects. 2023 Dec 8;14(12):936. doi: 10.3390/insects14120936. Insects. 2023. PMID: 38132609 Free PMC article.
-
De novo Transcriptomic analysis to unveil the deltamethrin induced resistance mechanisms in Callosobruchus chinensis (L.).Sci Rep. 2025 Feb 12;15(1):5163. doi: 10.1038/s41598-025-89466-3. Sci Rep. 2025. PMID: 39939732 Free PMC article.
-
Trichomes and unique gene expression confer insect herbivory resistance in Vitis labrusca grapevines.BMC Plant Biol. 2024 Jun 27;24(1):609. doi: 10.1186/s12870-024-05260-9. BMC Plant Biol. 2024. PMID: 38926877 Free PMC article.
References
LinkOut - more resources
Full Text Sources

