Identification of afatinib-associated ADH1B and potential small-molecule drugs targeting ADH1B for hepatocellular carcinoma
- PMID: 37229243
- PMCID: PMC10203513
- DOI: 10.3389/fphar.2023.1166454
Identification of afatinib-associated ADH1B and potential small-molecule drugs targeting ADH1B for hepatocellular carcinoma
Abstract
Background: Afatinib is an irreversible epidermal growth factor receptor tyrosine kinase inhibitor, and it plays a role in hepatocellular carcinoma (LIHC). This study aimed to screen a key gene associated with afatinib and identify its potential candidate drugs. Methods: We screened afatinib-associated differential expressed genes based on transcriptomic data of LIHC patients from The Cancer Genome Atlas, Gene Expression Omnibus, and the Hepatocellular Carcinoma Database (HCCDB). By using the Genomics of Drug Sensitivity in Cancer 2 database, we determined candidate genes using analysis of the correlation between differential genes and half-maximal inhibitory concentration. Survival analysis of candidate genes was performed in the TCGA dataset and validated in HCCDB18 and GSE14520 datasets. Immune characteristic analysis identified a key gene, and we found potential candidate drugs using CellMiner. We also evaluated the correlation between the expression of ADH1B and its methylation level. Furthermore, Western blot analysis was performed to validate the expression of ADH1B in normal hepatocytes LO2 and LIHC cell line HepG2. Results: We screened eight potential candidate genes (ASPM, CDK4, PTMA, TAT, ADH1B, ANXA10, OGDHL, and PON1) associated with afatinib. Patients with higher ASPM, CDK4, PTMA, and TAT exhibited poor prognosis, while those with lower ADH1B, ANXA10, OGDHL, and PON1 had unfavorable prognosis. Next, ADH1B was identified as a key gene negatively correlated with the immune score. The expression of ADH1B was distinctly downregulated in tumor tissues of pan-cancer. The expression of ADH1B was negatively correlated with ADH1B methylation. Small-molecule drugs panobinostat, oxaliplatin, ixabepilone, and seliciclib were significantly associated with ADH1B. The protein level of ADH1B was significantly downregulated in HepG2 cells compared with LO2 cells. Conclusion: Our study provides ADH1B as a key afatinib-related gene, which is associated with the immune microenvironment and can be used to predict the prognosis of LIHC. It is also a potential target of candidate drugs, sharing a promising approach to the development of novel drugs for the treatment of LIHC.
Keywords: ADH1B; CellMiner; afatinib; hepatocellular carcinoma; methylation; small-molecule drugs.
Copyright © 2023 Zhou, Yu, Huang, Zhao, He, Cui, Pan and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



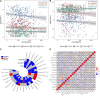

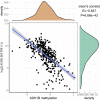


References
-
- Alsfouk A. A., Alshibl H. M., Altwaijry N. A., Alsfouk B. A., Al-Abdullah E. S. (2021). Synthesis and biological evaluation of seliciclib derivatives as potent and selective CDK9 inhibitors for prostate cancer therapy. Monatsh. für Chemie-Chemical Mon. 152, 109–120. 10.1007/s00706-020-02727-x - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

