Fragment Merging Using a Graph Database Samples Different Catalogue Space than Similarity Search
- PMID: 37229647
- PMCID: PMC10268959
- DOI: 10.1021/acs.jcim.3c00276
Fragment Merging Using a Graph Database Samples Different Catalogue Space than Similarity Search
Abstract
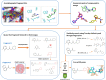
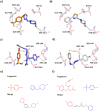
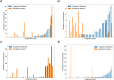
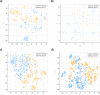
Fragment merging is a promising approach to progressing fragments directly to on-scale potency: each designed compound incorporates the structural motifs of overlapping fragments in a way that ensures compounds recapitulate multiple high-quality interactions. Searching commercial catalogues provides one useful way to quickly and cheaply identify such merges and circumvents the challenge of synthetic accessibility, provided they can be readily identified. Here, we demonstrate that the Fragment Network, a graph database that provides a novel way to explore the chemical space surrounding fragment hits, is well-suited to this challenge. We use an iteration of the database containing >120 million catalogue compounds to find fragment merges for four crystallographic screening campaigns and contrast the results with a traditional fingerprint-based similarity search. The two approaches identify complementary sets of merges that recapitulate the observed fragment-protein interactions but lie in different regions of chemical space. We further show our methodology is an effective route to achieving on-scale potency by retrospective analyses for two different targets; in analyses of public COVID Moonshot and Mycobacterium tuberculosis EthR inhibitors, potential inhibitors with micromolar IC50 values were identified. This work demonstrates the use of the Fragment Network to increase the yield of fragment merges beyond that of a classical catalogue search.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Davis B. J.; Roughley S. D. In Platform Technologies in Drug Discovery and Validation; Goodnow R. A., Ed.; Annual Reports in Medicinal Chemistry series; Academic Press, 2017; Vol. 50; pp 371–439.
-
- Keserű G. M.; Erlanson D. A.; Ferenczy G. G.; Hann M. M.; Murray C. W.; Pickett S. D. Design principles for fragment libraries: maximizing the value of learnings from pharma fragment-based drug discovery (FBDD) programs for use in academia. J. Med. Chem. 2016, 59, 8189–8206. 10.1021/acs.jmedchem.6b00197. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

