Simple and efficient differentiation of human iPSCs into contractible skeletal muscles for muscular disease modeling
- PMID: 37231024
- PMCID: PMC10213064
- DOI: 10.1038/s41598-023-34445-9
Simple and efficient differentiation of human iPSCs into contractible skeletal muscles for muscular disease modeling
Abstract
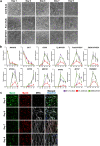
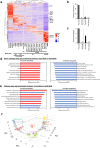
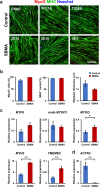
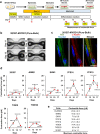
Pathophysiological analysis and drug discovery targeting human diseases require disease models that suitably recapitulate patient pathology. Disease-specific human induced pluripotent stem cells (hiPSCs) differentiated into affected cell types can potentially recapitulate disease pathology more accurately than existing disease models. Such successful modeling of muscular diseases requires efficient differentiation of hiPSCs into skeletal muscles. hiPSCs transduced with doxycycline-inducible MYOD1 (MYOD1-hiPSCs) have been widely used; however, they require time- and labor-consuming clonal selection, and clonal variations must be overcome. Moreover, their functionality should be carefully examined. Here, we demonstrated that bulk MYOD1-hiPSCs established with puromycin selection rather than G418 selection showed rapid and highly efficient differentiation. Interestingly, bulk MYOD1-hiPSCs exhibited average differentiation properties of clonally established MYOD1-hiPSCs, suggesting that it is possible to minimize clonal variations. Moreover, disease-specific hiPSCs of spinal bulbar muscular atrophy (SBMA) could be efficiently differentiated via this method into skeletal muscle that showed disease phenotypes, suggesting the applicability of this method for disease analysis. Finally, three-dimensional muscle tissues were fabricated from bulk MYOD1-hiPSCs, which exhibited contractile force upon electrical stimulation, indicating their functionality. Thus, our bulk differentiation requires less time and labor than existing methods, efficiently generates contractible skeletal muscles, and may facilitate the generation of muscular disease models.
© 2023. The Author(s).
Conflict of interest statement
HO is a paid member of the Scientific Advisory Board of SanBio Co., Ltd., and YO is a scientific advisor of Kohjin Bio Co., Ltd. The other authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

