The impact of rare protein coding genetic variation on adult cognitive function
- PMID: 37231097
- PMCID: PMC10260403
- DOI: 10.1038/s41588-023-01398-8
The impact of rare protein coding genetic variation on adult cognitive function
Abstract
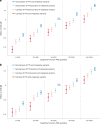
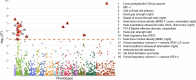
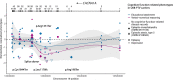
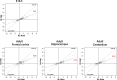
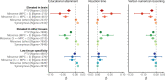
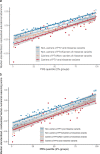
Compelling evidence suggests that human cognitive function is strongly influenced by genetics. Here, we conduct a large-scale exome study to examine whether rare protein-coding variants impact cognitive function in the adult population (n = 485,930). We identify eight genes (ADGRB2, KDM5B, GIGYF1, ANKRD12, SLC8A1, RC3H2, CACNA1A and BCAS3) that are associated with adult cognitive function through rare coding variants with large effects. Rare genetic architecture for cognitive function partially overlaps with that of neurodevelopmental disorders. In the case of KDM5B we show how the genetic dosage of one of these genes may determine the variability of cognitive, behavioral and molecular traits in mice and humans. We further provide evidence that rare and common variants overlap in association signals and contribute additively to cognitive function. Our study introduces the relevance of rare coding variants for cognitive function and unveils high-impact monogenic contributions to how cognitive function is distributed in the normal adult population.
© 2023. The Author(s).
Conflict of interest statement
C.-Y.C., T.F., E.A.T., H.R. and members of the Biogen Biobank team are employees of Biogen. R.T. is an employee of Dewpoint Therapeutics. J.Z.L. is an employee of GSK. M.E.H. is a cofounder, shareholder and nonexecutive director of Congenica, and an advisor to AstraZeneca. The other authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

