Divergent metabolic programmes control two populations of MAIT cells that protect the lung
- PMID: 37231163
- PMCID: PMC10264248
- DOI: 10.1038/s41556-023-01152-6
Divergent metabolic programmes control two populations of MAIT cells that protect the lung
Abstract
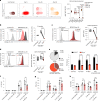
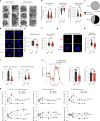
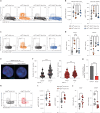
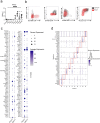
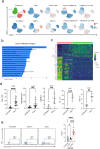
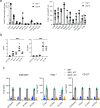
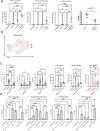
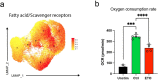
Although mucosal-associated invariant T (MAIT) cells provide rapid, innate-like responses, they are not pre-set, and memory-like responses have been described for MAIT cells following infections. The importance of metabolism for controlling these responses, however, is unknown. Here, following pulmonary immunization with a Salmonella vaccine strain, mouse MAIT cells expanded as separate CD127-Klrg1+ and CD127+Klrg1- antigen-adapted populations that differed in terms of their transcriptome, function and localization in lung tissue. These populations remained altered from steady state for months as stable, separate MAIT cell lineages with enhanced effector programmes and divergent metabolism. CD127+ MAIT cells engaged in an energetic, mitochondrial metabolic programme, which was critical for their maintenance and IL-17A synthesis. This programme was supported by high fatty acid uptake and mitochondrial oxidation and relied on highly polarized mitochondria and autophagy. After vaccination, CD127+ MAIT cells protected mice against Streptococcus pneumoniae infection. In contrast, Klrg1+ MAIT cells had dormant but ready-to-respond mitochondria and depended instead on Hif1a-driven glycolysis to survive and produce IFN-γ. They responded antigen independently and participated in protection from influenza virus. These metabolic dependencies may enable tuning of memory-like MAIT cell responses for vaccination and immunotherapies.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures









Comment in
-
Host protection by two memory-like MAIT subsets.Nat Cell Biol. 2023 Jun;25(6):802-804. doi: 10.1038/s41556-023-01149-1. Nat Cell Biol. 2023. PMID: 37231162 No abstract available.

