Therapeutic targeting of P2X4 receptor and mitochondrial metabolism in clear cell renal carcinoma models
- PMID: 37231503
- PMCID: PMC10214673
- DOI: 10.1186/s13046-023-02713-1
Therapeutic targeting of P2X4 receptor and mitochondrial metabolism in clear cell renal carcinoma models
Abstract
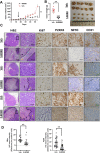
Background: Clear cell renal cell carcinoma (ccRCC) is the most common subtype of renal cancer. Large-scale metabolomic data have associated metabolic alterations with the pathogenesis and progression of renal carcinoma and have correlated mitochondrial activity with poor survival in a subset of patients. The aim of this study was to determine whether targeting mitochondria-lysosome interaction could be a novel therapeutic approach using patient-derived organoids as avatar for drug response.
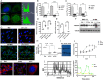
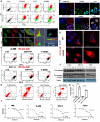
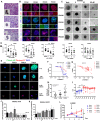
Methods: RNAseq data analysis and immunohistochemistry were used to show overexpression of Purinergic receptor 4 (P2XR4) in clear cell carcinomas. Seahorse experiments, immunofluorescence and fluorescence cell sorting were used to demonstrate that P2XR4 regulates mitochondrial activity and the balance of radical oxygen species. Pharmacological inhibitors and genetic silencing promoted lysosomal damage, calcium overload in mitochondria and cell death via both necrosis and apoptosis. Finally, we established patient-derived organoids and murine xenograft models to investigate the antitumor effect of P2XR4 inhibition using imaging drug screening, viability assay and immunohistochemistry.
Results: Our data suggest that oxo-phosphorylation is the main source of tumor-derived ATP in a subset of ccRCC cells expressing P2XR4, which exerts a critical impact on tumor energy metabolism and mitochondrial activity. Prolonged mitochondrial failure induced by pharmacological inhibition or P2XR4 silencing was associated with increased oxygen radical species, changes in mitochondrial permeability (i.e., opening of the transition pore complex, dissipation of membrane potential, and calcium overload). Interestingly, higher mitochondrial activity in patient derived organoids was associated with greater sensitivity to P2XR4 inhibition and tumor reduction in a xenograft model.
Conclusion: Overall, our results suggest that the perturbed balance between lysosomal integrity and mitochondrial activity induced by P2XR4 inhibition may represent a new therapeutic strategy for a subset of patients with renal carcinoma and that individualized organoids may be help to predict drug efficacy.
Keywords: Drug screening; Lysosomes; Mitochondria; Organoids; Purinergic receptors; Renal carcinoma.
© 2023. The Author(s).
Conflict of interest statement
None.
Figures