Out of Line or Altered States? Neural Progenitors as a Target in a Polygenic Neurodevelopmental Disorder
- PMID: 37231803
- PMCID: PMC12184714
- DOI: 10.1159/000530898
Out of Line or Altered States? Neural Progenitors as a Target in a Polygenic Neurodevelopmental Disorder
Abstract
The genesis of a mature complement of neurons is thought to require, at least in part, precursor cell lineages in which neural progenitors have distinct identities recognized by exclusive expression of one or a few molecular markers. Nevertheless, limited progenitor types distinguished by specific markers and lineal progression through such subclasses cannot easily yield the magnitude of neuronal diversity in most regions of the nervous system. The late Verne Caviness, to whom this edition of Developmental Neuroscience is dedicated, recognized this mismatch. In his pioneering work on the histogenesis of the cerebral cortex, he acknowledged the additional flexibility required to generate multiple classes of cortical projection and interneurons. This flexibility may be accomplished by establishing cell states in which levels rather than binary expression or repression of individual genes vary across each progenitor's shared transcriptome. Such states may reflect local, stochastic signaling via soluble factors or coincidence of cell surface ligand/receptor pairs in subsets of neighboring progenitors. This probabilistic, rather than determined, signaling could modify transcription levels via multiple pathways within an apparently uniform population of progenitors. Progenitor states, therefore, rather than lineal relationships between types may underlie the generation of neuronal diversity in most regions of the nervous system. Moreover, mechanisms that influence variation required for flexible progenitor states may be targets for pathological changes in a broad range of neurodevelopmental disorders, especially those with polygenic origins.
Keywords: 22q11 deletion syndrome; Animal model; Cell lineage; Cerebral cortex; Neural development; Neurogenesis.
© 2023 The Author(s). Published by S. Karger AG, Basel.
Conflict of interest statement
Conflict of Interest Statement
None of the authors have conflicts of interest to declare.
Figures




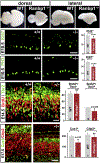

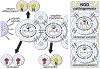
References
-
- Rakic P, Caviness VS Jr. Cortical development: view from neurological mutants two decades later. Neuron. 1995. Jun;14(6):1101–4. - PubMed
-
- Caviness VS Jr, Rakic P. Mechanisms of cortical development: a view from mutations in mice. Annu Rev Neurosci. 1978;1:297–326. - PubMed
-
- Herrup K. Role of staggerer gene in determining cell number in cerebellar cortex. I. Granule cell death is an indirect consequence of staggerer gene action. Brain Res. 1983. Dec;313(2):267–74. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

