Proximity labelling identifies pro-migratory endocytic recycling cargo and machinery of the Rab4 and Rab11 families
- PMID: 37232246
- PMCID: PMC10323252
- DOI: 10.1242/jcs.260468
Proximity labelling identifies pro-migratory endocytic recycling cargo and machinery of the Rab4 and Rab11 families
Abstract
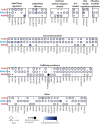
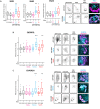
Endocytic recycling controls the return of internalised cargoes to the plasma membrane to coordinate their positioning, availability and downstream signalling. The Rab4 and Rab11 small GTPase families regulate distinct recycling routes, broadly classified as fast recycling from early endosomes (Rab4) and slow recycling from perinuclear recycling endosomes (Rab11), and both routes handle a broad range of overlapping cargoes to regulate cell behaviour. We adopted a proximity labelling approach, BioID, to identify and compare the protein complexes recruited by Rab4a, Rab11a and Rab25 (a Rab11 family member implicated in cancer aggressiveness), revealing statistically robust protein-protein interaction networks of both new and well-characterised cargoes and trafficking machinery in migratory cancer cells. Gene ontological analysis of these interconnected networks revealed that these endocytic recycling pathways are intrinsically connected to cell motility and cell adhesion. Using a knock-sideways relocalisation approach, we were further able to confirm novel links between Rab11, Rab25 and the ESCPE-1 and retromer multiprotein sorting complexes, and identify new endocytic recycling machinery associated with Rab4, Rab11 and Rab25 that regulates cancer cell migration in the 3D matrix.
Keywords: Cell migration; Endocytic recycling; Rab11; Rab25; Rab4.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures






References
-
- Caswell, P. T., Spence, H. J., Parsons, M., White, D. P., Clark, K., Cheng, K. W., Mills, G. B., Humphries, M. J., Messent, A. J., Anderson, K. I.et al. (2007). Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell 13, 496-510. 10.1016/j.devcel.2007.08.012 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

