Chemo-proteomics in antimalarial target identification and engagement
- PMID: 37232495
- PMCID: PMC10947479
- DOI: 10.1002/med.21975
Chemo-proteomics in antimalarial target identification and engagement
Abstract
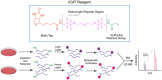
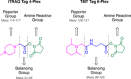
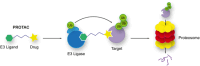
Humans have lived in tenuous battle with malaria over millennia. Today, while much of the world is free of the disease, areas of South America, Asia, and Africa still wage this war with substantial impacts on their social and economic development. The threat of widespread resistance to all currently available antimalarial therapies continues to raise concern. Therefore, it is imperative that novel antimalarial chemotypes be developed to populate the pipeline going forward. Phenotypic screening has been responsible for the majority of the new chemotypes emerging in the past few decades. However, this can result in limited information on the molecular target of these compounds which may serve as an unknown variable complicating their progression into clinical development. Target identification and validation is a process that incorporates techniques from a range of different disciplines. Chemical biology and more specifically chemo-proteomics have been heavily utilized for this purpose. This review provides an in-depth summary of the application of chemo-proteomics in antimalarial development. Here we focus particularly on the methodology, practicalities, merits, and limitations of designing these experiments. Together this provides learnings on the future use of chemo-proteomics in antimalarial development.
Keywords: antimalarial; chemical probe; malaria; target engagement; target identification.
© 2023 The Authors. Medicinal Research Reviews published by Wiley Periodicals LLC.
Figures
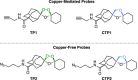
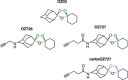
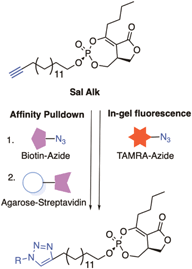
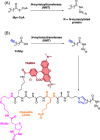
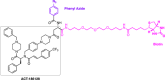
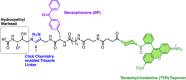
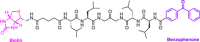
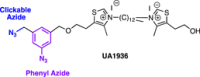
Similar articles
-
Efforts Aimed To Reduce Attrition in Antimalarial Drug Discovery: A Systematic Evaluation of the Current Antimalarial Targets Portfolio.ACS Infect Dis. 2018 Apr 13;4(4):568-576. doi: 10.1021/acsinfecdis.7b00211. Epub 2018 Jan 24. ACS Infect Dis. 2018. PMID: 29320160
-
[Combined antimalarial therapy using artemisinin].Parassitologia. 2004 Jun;46(1-2):85-7. Parassitologia. 2004. PMID: 15305693 Review. Italian.
-
The Development Process for Discovery and Clinical Advancement of Modern Antimalarials.J Med Chem. 2019 Dec 12;62(23):10526-10562. doi: 10.1021/acs.jmedchem.9b00761. Epub 2019 Aug 20. J Med Chem. 2019. PMID: 31385706 Review.
-
Spiral molecules with antimalarial activities: A review.Eur J Med Chem. 2022 Jul 5;237:114361. doi: 10.1016/j.ejmech.2022.114361. Epub 2022 Apr 8. Eur J Med Chem. 2022. PMID: 35461019 Review.
-
Medicinal Chemistry and Target Identification of Synthetic Clinical and Advanced Preclinical Antimalarial Candidates (2000 - 2022).Curr Top Med Chem. 2023;23(3):227-247. doi: 10.2174/1568026623666221220140526. Curr Top Med Chem. 2023. PMID: 36545719 Review.
Cited by
-
Evaluating Biocompatibility: From Classical Techniques to State-of-the-Art Functional Proteomics.Nanomaterials (Basel). 2025 Jul 3;15(13):1032. doi: 10.3390/nano15131032. Nanomaterials (Basel). 2025. PMID: 40648740 Free PMC article. Review.
-
Leveraging the Aggregated Protein Dye YAT2150 for Malaria Chemotherapy.Pharmaceutics. 2024 Sep 30;16(10):1290. doi: 10.3390/pharmaceutics16101290. Pharmaceutics. 2024. PMID: 39458619 Free PMC article.
-
Investigating Antiprotozoal Chemotherapies with Novel Proteomic Tools-Chances and Limitations: A Critical Review.Int J Mol Sci. 2024 Jun 24;25(13):6903. doi: 10.3390/ijms25136903. Int J Mol Sci. 2024. PMID: 39000012 Free PMC article. Review.
-
Eliminating malaria transmission requires targeting immature and mature gametocytes through lipoidal uptake of antimalarials.Nat Commun. 2024 Nov 15;15(1):9896. doi: 10.1038/s41467-024-54144-x. Nat Commun. 2024. PMID: 39548094 Free PMC article.
-
Chemoproteomics validates selective targeting of Plasmodium M1 alanyl aminopeptidase as an antimalarial strategy.Elife. 2024 Jul 8;13:RP92990. doi: 10.7554/eLife.92990. Elife. 2024. PMID: 38976500 Free PMC article.
References
-
- World Health Organization . World Malaria Report 2021. Geneva: World Health Organization; 2021.
-
- Korenromp EL, Williams BG, Gouws E, Dye C, Snow RW. Measurement of trends in childhood malaria mortality in Africa: an assessment of progress toward targets based on verbal autopsy. Lancet Infect Dis. 2003;3(6):349‐358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

