Identification of potential Indonesian marine invertebrate bioactive compounds as TMPRSS2 and SARS-CoV-2 Omicron spike protein inhibitors through computational screening
- PMID: 37234226
- PMCID: PMC10186851
- DOI: 10.1016/j.arabjc.2023.104984
Identification of potential Indonesian marine invertebrate bioactive compounds as TMPRSS2 and SARS-CoV-2 Omicron spike protein inhibitors through computational screening
Abstract
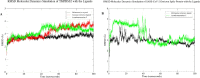
The coronavirus pandemic led to the announcement of a worldwide health emergency. The SARS-CoV-2 Omicron variant, which swiftly spread worldwide, has fueled existing challenges. Appropriate medication is necessary to avoid severe SARS-CoV-2 disease. The human TMPRSS2 and SARS-CoV-2 Omicron spike protein, which are required for viral entry into the host phase, were identified as the target proteins through computational screening. Structure-based virtual screening; molecular docking; absorption, distribution, metabolism, excretion, and toxicity (ADMET) analysis; and molecular dynamics simulation were the methods applied for TMPRSS2 and spike protein inhibitors. Bioactive marine invertebrates from Indonesia were employed as test ligands. Camostat and nafamostat (co-crystal) were utilized as reference ligands against TMPRSS2, whereas mefloquine was used as a reference ligand against spike protein. Following a molecular docking and dynamics simulation, we found that acanthomanzamine C has remarkable effectiveness against TMPRSS2 and spike protein. Compared to camostat (-8.25 kcal/mol), nafamostat (-6.52 kcal/mol), and mefloquine (-6.34 kcal/mol), acanthomanzamine C binds to TMPRSS2 and spike protein with binding energies of -9.75 kcal/mol and -9.19 kcal/mol, respectively. Furthermore, slight variances in the MD simulation demonstrated consistent binding to TMPRSS2 and spike protein after the initial 50 ns. These results are highly valuable in the search for a treatment for SARS-CoV-2 infection.
Keywords: ADMET; Marine invertebrates; Molecular docking; Molecular dynamics; SARS-CoV-2; TMPRSS2.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
Figures







References
-
- Breining P., Frølund A.L., Højen J.F., Gunst J.D., Staerke N.B., Saedder E., Cases-Thomas M., Little P., Nielsen L.P., Søgaard O.S., Kjolby M. Camostat Mesylate against SARS-CoV-2 and COVID-19—Rationale, Dosing and Safety. Basic Clin. Paharmacol. Toxicol. 2021;128(2):204–212. doi: 10.1111/bcpt.13533. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
