Phylogeny-Related Variations in Venomics: A Test in a Subset of Habu Snakes (Protobothrops)
- PMID: 37235384
- PMCID: PMC10223207
- DOI: 10.3390/toxins15050350
Phylogeny-Related Variations in Venomics: A Test in a Subset of Habu Snakes (Protobothrops)
Abstract
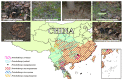
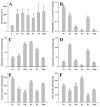
We conducted a comparative analysis to unveil the divergence among venoms from a subset of Old World habu snakes (Protobothrops) in terms of venomic profiles and toxicological and enzymatic activities. A total of 14 protein families were identified in the venoms from these habu snakes, and 11 of them were shared among these venoms. The venoms of five adult habu snakes were overwhelmingly dominated by SVMP (32.56 ± 13.94%), PLA2 (22.93 ± 9.26%), and SVSP (16.27 ± 4.79%), with a total abundance of over 65%, while the subadult P. mangshanensis had an extremely low abundance of PLA2 (1.23%) but a high abundance of CTL (51.47%), followed by SVMP (22.06%) and SVSP (10.90%). Apparent interspecific variations in lethality and enzymatic activities were also explored in habu snake venoms, but no variations in myotoxicity were found. Except for SVSP, the resemblance of the relatives within Protobothrops in other venom traits was estimated to deviate from Brownian motion evolution based on phylogenetic signals. A comparative analysis further validated that the degree of covariation between phylogeny and venom variation is evolutionarily labile and varies among clades of closely related snakes. Our findings indicate a high level of interspecific variation in the venom proteomes of habu snakes, both in the presence or absence and the relative abundance of venom protein families, and that these venoms might have evolved under a combination of adaptive and neutral mechanisms.
Keywords: Protobothrops; biochemical activity; interspecific variation; phylogenetic signal; venom proteome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

