Shaping Nanobodies and Intrabodies against Proteoforms
- PMID: 37235478
- PMCID: PMC10269583
- DOI: 10.1021/acs.analchem.3c00958
Shaping Nanobodies and Intrabodies against Proteoforms
Abstract
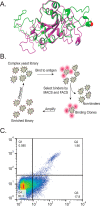
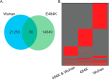
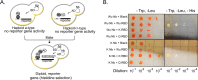
Proteoforms expand genomic diversity and direct developmental processes. While high-resolution mass spectrometry has accelerated characterization of proteoforms, molecular techniques working to bind and disrupt the function of specific proteoforms have lagged behind. In this study, we worked to develop intrabodies capable of binding specific proteoforms. We employed a synthetic camelid nanobody library expressed in yeast to identify nanobody binders of different SARS-CoV-2 receptor binding domain (RBD) proteoforms. Importantly, employment of the positive and negative selection mechanisms inherent to the synthetic system allowed for amplification of nanobody-expressing yeast that bind to the original (Wuhan strain RBD) but not the E484 K (Beta variant) mutation. Nanobodies raised against specific RBD proteoforms were validated by yeast-2-hybrid analysis and sequence comparisons. These results provide a framework for development of nanobodies and intrabodies that target proteoforms.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Adams L. M.; DeHart C. J.; Drown B. S.; Anderson L. C.; Bocik W.; Boja E. S.; Hiltke T. M.; Hendrickson C. L.; Rodriguez H.; Caldwell M.; Vafabakhsh R.; Kelleher N. L. Mapping the KRAS proteoform landscape in colorectal cancer identifies truncated KRAS4B that decreases MAPK signaling. J. Biol. Chem. 2023, 299 (1), 102768.10.1016/j.jbc.2022.102768. - DOI - PMC - PubMed
-
- Ntai I.; Fornelli L.; DeHart C. J.; Hutton J. E.; Doubleday P. F.; LeDuc R. D.; van Nispen A. J.; Fellers R. T.; Whiteley G.; Boja E. S.; Rodriguez H.; Kelleher N. L. Precise characterization of KRAS4b proteoforms in human colorectal cells and tumors reveals mutation/modification cross-talk. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (16), 4140–4145. 10.1073/pnas.1716122115. - DOI - PMC - PubMed
-
- Melani R. D.; Gerbasi V. R.; Anderson L. C.; Sikora J. W.; Toby T. K.; Hutton J. E.; Butcher D. S.; Negrao F.; Seckler H. S.; Srzentic K.; Fornelli L.; Camarillo J. M.; LeDuc R. D.; Cesnik A. J.; Lundberg E.; Greer J. B.; Fellers R. T.; Robey M. T.; DeHart C. J.; Forte E.; Hendrickson C. L.; Abbatiello S. E.; Thomas P. M.; Kokaji A. I.; Levitsky J.; Kelleher N. L. The Blood Proteoform Atlas: A reference map of proteoforms in human hematopoietic cells. Science 2022, 375 (6579), 411–418. 10.1126/science.aaz5284. - DOI - PMC - PubMed
-
- Toby T. K.; Abecassis M.; Kim K.; Thomas P. M.; Fellers R. T.; LeDuc R. D.; Kelleher N. L.; Demetris J.; Levitsky J. Proteoforms in Peripheral Blood Mononuclear Cells as Novel Rejection Biomarkers in Liver Transplant Recipients. Am. J. Transplant 2017, 17 (9), 2458–2467. 10.1111/ajt.14359. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

