Shape complexity in cluster analysis
- PMID: 37235568
- PMCID: PMC10218739
- DOI: 10.1371/journal.pone.0286312
Shape complexity in cluster analysis
Abstract
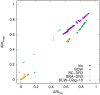
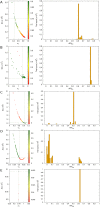
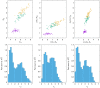
In cluster analysis, a common first step is to scale the data aiming to better partition them into clusters. Even though many different techniques have throughout many years been introduced to this end, it is probably fair to say that the workhorse in this preprocessing phase has been to divide the data by the standard deviation along each dimension. Like division by the standard deviation, the great majority of scaling techniques can be said to have roots in some sort of statistical take on the data. Here we explore the use of multidimensional shapes of data, aiming to obtain scaling factors for use prior to clustering by some method, like k-means, that makes explicit use of distances between samples. We borrow from the field of cosmology and related areas the recently introduced notion of shape complexity, which in the variant we use is a relatively simple, data-dependent nonlinear function that we show can be used to help with the determination of appropriate scaling factors. Focusing on what might be called "midrange" distances, we formulate a constrained nonlinear programming problem and use it to produce candidate scaling-factor sets that can be sifted on the basis of further considerations of the data, say via expert knowledge. We give results on some iconic data sets, highlighting the strengths and potential weaknesses of the new approach. These results are generally positive across all the data sets used.
Copyright: © 2023 Aguilar, Barbosa. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Milligan GW, Cooper MC. A study of standardization of variables in cluster analysis. J Classif. 1988;5:181–204. doi: 10.1007/BF01897163 - DOI
-
- Steinley D. Standardizing variables in k-means clustering. In: Banks D, McMorris FR, Arabie P, Gaul W, editors. Classification, Clustering, and Data Mining Applications. Berlin, Germany: Springer-Verlag; 2004. p. 53–60.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

