Differential Responses of the GLP-1 and GLP-2 Receptors to N-Terminal Modification of a Dual Agonist
- PMID: 37235770
- PMCID: PMC10335629
- DOI: 10.1021/jacs.3c01628
Differential Responses of the GLP-1 and GLP-2 Receptors to N-Terminal Modification of a Dual Agonist
Abstract
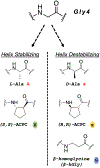
Class B1 G protein-coupled receptors (GPCRs), collectively, respond to a diverse repertoire of extracellular polypeptide agonists and transmit the encoded messages to cytosolic partners. To fulfill these tasks, these highly mobile receptors must interconvert among conformational states in response to agonists. We recently showed that conformational mobility in polypeptide agonists themselves plays a role in activation of one class B1 GPCR, the receptor for glucagon-like peptide-1 (GLP-1). Exchange between helical and nonhelical conformations near the N-termini of agonists bound to the GLP-1R was revealed to be critical for receptor activation. Here, we ask whether agonist conformational mobility plays a role in the activation of a related receptor, the GLP-2R. Using variants of the hormone GLP-2 and the designed clinical agonist glepaglutide (GLE), we find that the GLP-2R is quite tolerant of variations in α-helical propensity near the agonist N-terminus, which contrasts with signaling at the GLP-1R. A fully α-helical conformation of the bound agonist may be sufficient for GLP-2R signal transduction. GLE is a GLP-2R/GLP-1R dual agonist, and the GLE system therefore enables direct comparison of the responses of these two GPCRs to a single set of agonist variants. This comparison supports the conclusion that the GLP-1R and GLP-2R differ in their response to variations in helical propensity near the agonist N-terminus. The data offer a basis for development of new hormone analogues with distinctive and potentially useful activity profiles; for example, one of the GLE analogues is a potent agonist of the GLP-2R but also a potent antagonist of the GLP-1R, a novel form of polypharmacology.
Figures



Similar articles
-
Cryo-electron microscopy structure of the glucagon receptor with a dual-agonist peptide.J Biol Chem. 2020 Jul 10;295(28):9313-9325. doi: 10.1074/jbc.RA120.013793. Epub 2020 May 5. J Biol Chem. 2020. PMID: 32371397 Free PMC article.
-
Effects of Replacing a Central Glycine Residue in GLP-1 on Receptor Affinity and Signaling Profile.Chembiochem. 2023 Nov 2;24(21):e202300504. doi: 10.1002/cbic.202300504. Epub 2023 Sep 13. Chembiochem. 2023. PMID: 37624685 Free PMC article.
-
Differential Requirement of the Extracellular Domain in Activation of Class B G Protein-coupled Receptors.J Biol Chem. 2016 Jul 15;291(29):15119-30. doi: 10.1074/jbc.M116.726620. Epub 2016 May 13. J Biol Chem. 2016. PMID: 27226600 Free PMC article.
-
Insights into the structure and activation mechanism of some class B1 GPCR family members.Mol Biol Rep. 2024 Sep 6;51(1):966. doi: 10.1007/s11033-024-09876-w. Mol Biol Rep. 2024. PMID: 39240462 Review.
-
Glucagon-Like Peptide-1 and Its Class B G Protein-Coupled Receptors: A Long March to Therapeutic Successes.Pharmacol Rev. 2016 Oct;68(4):954-1013. doi: 10.1124/pr.115.011395. Pharmacol Rev. 2016. PMID: 27630114 Free PMC article. Review.
Cited by
-
Flexible fluorine-thiol displacement stapled peptides with enhanced membrane penetration for the estrogen receptor/coactivator interaction.J Biol Chem. 2024 Dec;300(12):107991. doi: 10.1016/j.jbc.2024.107991. Epub 2024 Nov 13. J Biol Chem. 2024. PMID: 39547512 Free PMC article.
-
Glepaglutide-Loaded Foam for the Induction of Mucosal Healing in the Treatment of Inflammatory Bowel Disease.Adv Healthc Mater. 2025 Mar;14(7):e2403497. doi: 10.1002/adhm.202403497. Epub 2025 Feb 5. Adv Healthc Mater. 2025. PMID: 39905897 Free PMC article.
-
Hormone Analogues with Unique Signaling Profiles from Replacement of α-Residue Triads with β/γ Diads.J Am Chem Soc. 2023 Sep 20;145(37):20539-20550. doi: 10.1021/jacs.3c06703. Epub 2023 Sep 11. J Am Chem Soc. 2023. PMID: 37697685 Free PMC article.
-
Prolonged signaling of backbone-modified glucagon-like peptide-1 analogues with diverse receptor trafficking.Proc Natl Acad Sci U S A. 2025 Apr 8;122(14):e2407574122. doi: 10.1073/pnas.2407574122. Epub 2025 Apr 1. Proc Natl Acad Sci U S A. 2025. PMID: 40168114
References
-
- Deganutti G; Liang YL; Zhang X; Khoshouei M; Clydesdale L; Belousoff MJ; Venugopal H; Truong TT; Glukhova A; Keller AN; Gregory KJ; Leach K; Christopoulos A; Danev R; Reynolds CA; Zhao P; Sexton PM; Wootten D Dynamics of GLP-1R Peptide Agonist Engagement Are Correlated with Kinetics of G Protein Activation. Nature Communications 2022, 13 (1), 1–18. 10.1038/s41467-021-27760-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

