Pre-mRNA splicing and its cotranscriptional connections
- PMID: 37236814
- PMCID: PMC10524715
- DOI: 10.1016/j.tig.2023.04.008
Pre-mRNA splicing and its cotranscriptional connections
Abstract
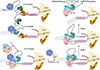
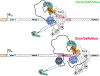
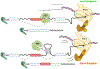
Transcription of eukaryotic genes by RNA polymerase II (Pol II) yields RNA precursors containing introns that must be spliced out and the flanking exons ligated together. Splicing is catalyzed by a dynamic ribonucleoprotein complex called the spliceosome. Recent evidence has shown that a large fraction of splicing occurs cotranscriptionally as the RNA chain is extruded from Pol II at speeds of up to 5 kb/minute. Splicing is more efficient when it is tethered to the transcription elongation complex, and this linkage permits functional coupling of splicing with transcription. We discuss recent progress that has uncovered a network of connections that link splicing to transcript elongation and other cotranscriptional RNA processing events.
Keywords: RNA pol II; U1 snRNP; exon definition; intron definition; nascent RNA folding; pre-mRNA splicing; transcription elongation.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no conflicts of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

