TRABID inhibition activates cGAS/STING-mediated anti-tumor immunity through mitosis and autophagy dysregulation
- PMID: 37237031
- PMCID: PMC10220035
- DOI: 10.1038/s41467-023-38784-z
TRABID inhibition activates cGAS/STING-mediated anti-tumor immunity through mitosis and autophagy dysregulation
Abstract
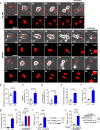
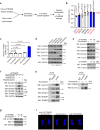
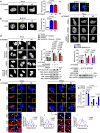
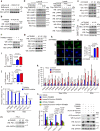
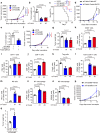
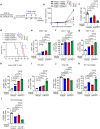
Activation of tumor-intrinsic innate immunity has been a major strategy for improving immunotherapy. Previously, we reported an autophagy-promoting function of the deubiquitinating enzyme TRABID. Here, we identify a critical role of TRABID in suppressing anti-tumor immunity. Mechanistically, TRABID is upregulated in mitosis and governs mitotic cell division by removing K29-linked polyubiquitin chain from Aurora B and Survivin, thereby stabilizing the entire chromosomal passenger complex. TRABID inhibition causes micronuclei through a combinatory defect in mitosis and autophagy and protects cGAS from autophagic degradation, thereby activating the cGAS/STING innate immunity pathway. Genetic or pharmacological inhibition of TRABID promotes anti-tumor immune surveillance and sensitizes tumors to anti-PD-1 therapy in preclinical cancer models in male mice. Clinically, TRABID expression in most solid cancer types correlates inversely with an interferon signature and infiltration of anti-tumor immune cells. Our study identifies a suppressive role of tumor-intrinsic TRABID in anti-tumor immunity and highlights TRABID as a promising target for sensitizing solid tumors to immunotherapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

