RNA ligase ribozymes with a small catalytic core
- PMID: 37237056
- PMCID: PMC10219994
- DOI: 10.1038/s41598-023-35584-9
RNA ligase ribozymes with a small catalytic core
Abstract
Catalytic RNAs, or ribozymes, catalyze diverse chemical reactions that could have sustained primordial life in the hypothetical RNA world. Many natural ribozymes and laboratory evolved ribozymes exhibit efficient catalysis mediated by elaborate catalytic cores within complex tertiary structures. However, such complex RNA structures and sequences are unlikely to have emerged by chance during the earliest phase of chemical evolution. Here, we explored simple and small ribozyme motifs capable of ligating two RNA fragments in a template-directed fashion (ligase ribozymes). One-round selection of small ligase ribozymes followed by deep sequencing revealed a ligase ribozyme motif comprising a three-nucleotide loop opposite to the ligation junction. The observed ligation was magnesium(II) dependent and appears to form a 2'-5' phosphodiester linkage. The fact that such a small RNA motif can function as a catalyst supports a scenario in which RNA or other primordial nucleic acids played a central role in chemical evolution of life.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
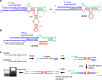


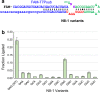
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

