Exploring the prognostic function of TMB-related prognostic signature in patients with colon cancer
- PMID: 37237274
- PMCID: PMC10214595
- DOI: 10.1186/s12920-023-01555-2
Exploring the prognostic function of TMB-related prognostic signature in patients with colon cancer
Abstract
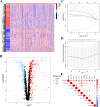
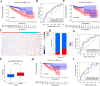
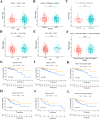
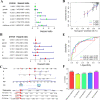
Tumor mutation burden (TMB) level is identified as a useful predictor in multiple tumors including colon adenocarcinoma (COAD). However, the function of TMB related genes has not been explored previously. In this study, we obtained patients' expression and clinical data from The Cancer Genome Atlas (TCGA) and the National Center for Biotechnology Information (NCBI). TMB genes were screened and subjected to differential expression analysis. Univariate Cox and LASSO analyses were utilized to construct the prognostic signature. The efficiency of the signature was tested by using a receiver operating characteristic (ROC) curve. A nomogram was further plotted to assess the overall survival (OS) time of patients with COAD. In addition, we compared the predictive performance of our signature with other four published signatures. Functional analyses indicated that patients in the low-risk group have obviously different enrichment of tumor related pathways and tumor infiltrating immune cells from that of high-risk patients. Our findings suggested that the ten genes' prognostic signature could exert undeniable prognostic functions in patients with COAD, which might provide significant clues for the development of personalized management of these patients.
Keywords: Colon cancer; Prognosis; Risk signature; Tumor mutation burden.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests” to this section.
Figures



Similar articles
-
Identification of a glycolysis- and lactate-related gene signature for predicting prognosis, immune microenvironment, and drug candidates in colon adenocarcinoma.Front Cell Dev Biol. 2022 Aug 23;10:971992. doi: 10.3389/fcell.2022.971992. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36081904 Free PMC article.
-
An Intratumor Heterogeneity-Related Signature for Predicting Prognosis, Immune Landscape, and Chemotherapy Response in Colon Adenocarcinoma.Front Med (Lausanne). 2022 Jul 7;9:925661. doi: 10.3389/fmed.2022.925661. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35872794 Free PMC article.
-
Lactate Metabolism-Associated lncRNA Pairs: A Prognostic Signature to Reveal the Immunological Landscape and Mediate Therapeutic Response in Patients With Colon Adenocarcinoma.Front Immunol. 2022 Jul 11;13:881359. doi: 10.3389/fimmu.2022.881359. eCollection 2022. Front Immunol. 2022. PMID: 35911752 Free PMC article.
-
Exploration of the relationship between tumor mutation burden and immune infiltrates in colon adenocarcinoma.Int J Med Sci. 2021 Jan 1;18(3):685-694. doi: 10.7150/ijms.51918. eCollection 2021. Int J Med Sci. 2021. PMID: 33437203 Free PMC article.
-
Construction of a novel choline metabolism-related signature to predict prognosis, immune landscape, and chemotherapy response in colon adenocarcinoma.Front Immunol. 2022 Nov 14;13:1038927. doi: 10.3389/fimmu.2022.1038927. eCollection 2022. Front Immunol. 2022. PMID: 36451813 Free PMC article.
Cited by
-
Aluminum Concentration Is Associated with Tumor Mutational Burden and the Expression of Immune Response Biomarkers in Colorectal Cancers.Int J Mol Sci. 2024 Dec 13;25(24):13388. doi: 10.3390/ijms252413388. Int J Mol Sci. 2024. PMID: 39769153 Free PMC article.
-
Comprehensive characterization of MCL-1 in patients with colorectal cancer: Expression, molecular profiles, and outcomes.Int J Cancer. 2025 Apr 15;156(8):1583-1593. doi: 10.1002/ijc.35304. Epub 2024 Dec 30. Int J Cancer. 2025. PMID: 39740007 Free PMC article.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A, Cancer Statistics. 2021. CA: a cancer journal for clinicians. 2021; 71(1):7–33. 10.3322/caac.21654. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

