The biology of SCUBE
- PMID: 37237303
- PMCID: PMC10214685
- DOI: 10.1186/s12929-023-00925-3
The biology of SCUBE
Abstract
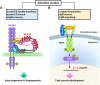
The SCUBE [Signal peptide-Complement C1r/C1s, Uegf, Bmp1 (CUB)-Epithelial growth factor domain-containing protein] family consists of three proteins in vertebrates, SCUBE1, 2 and 3, which are highly conserved in zebrafish, mice and humans. Each SCUBE gene encodes a polypeptide of approximately 1000 amino acids that is organized into five modular domains: (1) an N-terminal signal peptide sequence, (2) nine tandem epidermal growth factor (EGF)-like repeats, (3) a large spacer region, (4) three cysteine-rich (CR) motifs, and (5) a CUB domain at the C-terminus. Murine Scube genes are expressed individually or in combination during the development of various tissues, including those in the central nervous system and the axial skeleton. The cDNAs of human SCUBE orthologs were originally cloned from vascular endothelial cells, but SCUBE expression has also been found in platelets, mammary ductal epithelium and osteoblasts. Both soluble and membrane-associated SCUBEs have been shown to play important roles in physiology and pathology. For instance, upregulation of SCUBEs has been reported in acute myeloid leukemia, breast cancer and lung cancer. In addition, soluble SCUBE1 is released from activated platelets and can be used as a clinical biomarker for acute coronary syndrome and ischemic stroke. Soluble SCUBE2 enhances distal signaling by facilitating the secretion of dual-lipidated hedgehog from nearby ligand-producing cells in a paracrine manner. Interestingly, the spacer regions and CR motifs can increase or enable SCUBE binding to cell surfaces via electrostatic or glycan-lectin interactions. As such, membrane-associated SCUBEs can function as coreceptors that enhance the signaling activity of various serine/threonine kinase or tyrosine kinase receptors. For example, membrane-associated SCUBE3 functions as a coreceptor that promotes signaling in bone morphogenesis. In humans, SCUBE3 mutations are linked to abnormalities in growth and differentiation of both bones and teeth. In addition to studies on human SCUBE function, experimental results from genetically modified mouse models have yielded important insights in the field of systems biology. In this review, we highlight novel molecular discoveries and critical directions for future research on SCUBE proteins in the context of cancer, skeletal disease and cardiovascular disease.
Keywords: Biomarker; Coreceptor; Endothelial cells; SCUBE; Signal transduction.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Wu BT, Su YH, Tsai MT, Wasserman SM, Topper JN, Yang RB. A novel secreted, cell-surface glycoprotein containing multiple epidermal growth factor-like repeats and one CUB domain is highly expressed in primary osteoblasts and bones. J Biol Chem. 2004;279(36):37485–37490. doi: 10.1074/jbc.M405912200. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

