scSemiAAE: a semi-supervised clustering model for single-cell RNA-seq data
- PMID: 37237310
- PMCID: PMC10214737
- DOI: 10.1186/s12859-023-05339-4
scSemiAAE: a semi-supervised clustering model for single-cell RNA-seq data
Abstract
Background: Single-cell RNA sequencing (scRNA-seq) strives to capture cellular diversity with higher resolution than bulk RNA sequencing. Clustering analysis is critical to transcriptome research as it allows for further identification and discovery of new cell types. Unsupervised clustering cannot integrate prior knowledge where relevant information is widely available. Purely unsupervised clustering algorithms may not yield biologically interpretable clusters when confronted with the high dimensionality of scRNA-seq data and frequent dropout events, which makes identification of cell types more challenging.
Results: We propose scSemiAAE, a semi-supervised clustering model for scRNA sequence analysis using deep generative neural networks. Specifically, scSemiAAE carefully designs a ZINB adversarial autoencoder-based architecture that inherently integrates adversarial training and semi-supervised modules in the latent space. In a series of experiments on scRNA-seq datasets spanning thousands to tens of thousands of cells, scSemiAAE can significantly improve clustering performance compared to dozens of unsupervised and semi-supervised algorithms, promoting clustering and interpretability of downstream analyses.
Conclusion: scSemiAAE is a Python-based algorithm implemented on the VSCode platform that provides efficient visualization, clustering, and cell type assignment for scRNA-seq data. The tool is available from https://github.com/WHang98/scSemiAAE .
Keywords: Adversarial autoencoder; Clustering; Deep learning; Semi-supervised; scRNA-seq.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

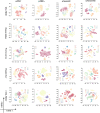
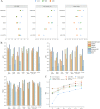
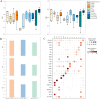
Similar articles
-
Deep structural clustering for single-cell RNA-seq data jointly through autoencoder and graph neural network.Brief Bioinform. 2022 Mar 10;23(2):bbac018. doi: 10.1093/bib/bbac018. Brief Bioinform. 2022. PMID: 35172334
-
scCNC: a method based on capsule network for clustering scRNA-seq data.Bioinformatics. 2022 Aug 2;38(15):3703-3709. doi: 10.1093/bioinformatics/btac393. Bioinformatics. 2022. PMID: 35699473
-
Graph contrastive learning as a versatile foundation for advanced scRNA-seq data analysis.Brief Bioinform. 2024 Sep 23;25(6):bbae558. doi: 10.1093/bib/bbae558. Brief Bioinform. 2024. PMID: 39487083 Free PMC article.
-
Machine learning and statistical methods for clustering single-cell RNA-sequencing data.Brief Bioinform. 2020 Jul 15;21(4):1209-1223. doi: 10.1093/bib/bbz063. Brief Bioinform. 2020. PMID: 31243426 Review.
-
Supervised application of internal validation measures to benchmark dimensionality reduction methods in scRNA-seq data.Brief Bioinform. 2021 Nov 5;22(6):bbab304. doi: 10.1093/bib/bbab304. Brief Bioinform. 2021. PMID: 34374742 Review.
Cited by
-
scTPC: a novel semisupervised deep clustering model for scRNA-seq data.Bioinformatics. 2024 May 2;40(5):btae293. doi: 10.1093/bioinformatics/btae293. Bioinformatics. 2024. PMID: 38684178 Free PMC article.
-
A robust multi-scale clustering framework for single-cell RNA-seq data analysis.Sci Rep. 2025 May 27;15(1):18543. doi: 10.1038/s41598-025-03603-6. Sci Rep. 2025. PMID: 40425750 Free PMC article.
References
-
- Petegrosso R, Li Z, Kuang R. Machine learning and statistical methods for clustering single-cell RNA-sequencing data. Brief Bioinform. 2020;21(4):1209–1223. - PubMed
-
- Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16(3):133–145. - PubMed
-
- Kiselev VY, Andrews TS, Hemberg M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet. 2019;20(5):273–282. - PubMed
-
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58(4):610–620. - PubMed
-
- Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14(9):618–630. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

