Differences in Faecal Microbiome Taxonomy, Diversity and Functional Potential in a Bovine Cohort Experimentally Challenged with Mycobacterium avium subsp. paratuberculosis (MAP)
- PMID: 37238082
- PMCID: PMC10215331
- DOI: 10.3390/ani13101652
Differences in Faecal Microbiome Taxonomy, Diversity and Functional Potential in a Bovine Cohort Experimentally Challenged with Mycobacterium avium subsp. paratuberculosis (MAP)
Abstract
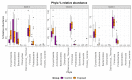
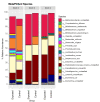
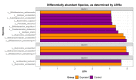
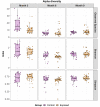
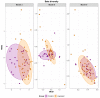
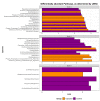
Mycobacterium avium subspecies paratuberculosis (MAP) is the causative agent of Johne's disease in ruminants, a chronic enteritis which results in emaciation and eventual loss of the animal. Recent advances in metagenomics have allowed a more in-depth study of complex microbiomes, including that of gastrointestinal tracts, and have the potential to provide insights into consequences of the exposure of an animal to MAP or other pathogens. This study aimed to investigate taxonomic diversity and compositional changes of the faecal microbiome of cattle experimentally challenged with MAP compared to an unexposed control group. Faecal swab samples were collected from a total of 55 animals [exposed group (n = 35) and a control group (n = 20)], across three time points (months 3, 6 and 9 post-inoculation). The composition and functional potential of the faecal microbiota differed across time and between the groups (p < 0.05), with the primary differences, from both a taxonomic and functional perspective, occurring at 3 months post inoculation. These included significant differences in the relative abundance of the genera Methanobrevibacter and Bifidobacterium and also of 11 other species (4 at a higher relative abundance in the exposed group and 7 at a higher relative abundance in the control group). Correlations were made between microbiome data and immunopathology measurements and it was noted that changes in the microbial composition correlated with miRNA-155, miR-146b and IFN-ɣ. In summary, this study illustrates the impact of exposure to MAP on the ruminant faecal microbiome with a number of species that may have relevance in veterinary medicine for tracking exposure to MAP.
Keywords: Johne’s disease; MAP; experimental model; immune function; microbiome.
Conflict of interest statement
The authors declare that there are no competing interests.
Figures
Similar articles
-
A long-term bacteriological and immunological study in Holstein-Friesian cattle experimentally infected with Mycobacterium avium subsp. paratuberculosis and necropsy culture results for Holstein-Friesian cattle, Merino sheep and Angora goats.Vet Microbiol. 2007 May 16;122(1-2):83-96. doi: 10.1016/j.vetmic.2006.12.030. Epub 2007 Jan 16. Vet Microbiol. 2007. PMID: 17289303
-
Faecal bacterial composition in dairy cows shedding Mycobacterium avium subsp. paratuberculosis in faeces in comparison with nonshedding cows.Can J Microbiol. 2016 Jun;62(6):538-41. doi: 10.1139/cjm-2015-0814. Epub 2016 Feb 29. Can J Microbiol. 2016. PMID: 27127920
-
Immunopathological changes and apparent recovery from infection revealed in cattle in an experimental model of Johne's disease using a lyophilised culture of Mycobacterium avium subspecies paratuberculosis.Vet Microbiol. 2018 Jun;219:53-62. doi: 10.1016/j.vetmic.2018.03.029. Epub 2018 Mar 29. Vet Microbiol. 2018. PMID: 29778205
-
Potential application of emerging diagnostic techniques to the diagnosis of bovine Johne's disease (paratuberculosis).Vet J. 2016 Mar;209:32-9. doi: 10.1016/j.tvjl.2015.10.033. Epub 2015 Oct 22. Vet J. 2016. PMID: 26831164 Review.
-
Facts, myths and hypotheses on the zoonotic nature of Mycobacterium avium subspecies paratuberculosis.Int J Med Microbiol. 2014 Oct;304(7):858-67. doi: 10.1016/j.ijmm.2014.07.006. Epub 2014 Jul 25. Int J Med Microbiol. 2014. PMID: 25128370 Review.
Cited by
-
Gut Microbiota Profiling as a Promising Tool to Detect Equine Inflammatory Bowel Disease (IBD).Animals (Basel). 2024 Aug 18;14(16):2396. doi: 10.3390/ani14162396. Animals (Basel). 2024. PMID: 39199930 Free PMC article.
References
-
- Rosa B.A., Supali T., Gankpala L., Djuardi Y., Sartono E., Zhou Y., Fischer K., Martin J., Tyagi R., Bolay F.K., et al. Differential Human Gut Microbiome Assemblages during Soil-Transmitted Helminth Infections in Indonesia and Liberia. Microbiome. 2018;6:33. doi: 10.1186/s40168-018-0416-5. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

