Ribosomal Protein uS5 and Friends: Protein-Protein Interactions Involved in Ribosome Assembly and Beyond
- PMID: 37238722
- PMCID: PMC10216425
- DOI: 10.3390/biom13050853
Ribosomal Protein uS5 and Friends: Protein-Protein Interactions Involved in Ribosome Assembly and Beyond
Abstract
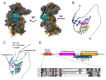
Ribosomal proteins are fundamental components of the ribosomes in all living cells. The ribosomal protein uS5 (Rps2) is a stable component of the small ribosomal subunit within all three domains of life. In addition to its interactions with proximal ribosomal proteins and rRNA inside the ribosome, uS5 has a surprisingly complex network of evolutionarily conserved non-ribosome-associated proteins. In this review, we focus on a set of four conserved uS5-associated proteins: the protein arginine methyltransferase 3 (PRMT3), the programmed cell death 2 (PDCD2) and its PDCD2-like (PDCD2L) paralog, and the zinc finger protein, ZNF277. We discuss recent work that presents PDCD2 and homologs as a dedicated uS5 chaperone and PDCD2L as a potential adaptor protein for the nuclear export of pre-40S subunits. Although the functional significance of the PRMT3-uS5 and ZNF277-uS5 interactions remain elusive, we reflect on the potential roles of uS5 arginine methylation by PRMT3 and on data indicating that ZNF277 and PRMT3 compete for uS5 binding. Together, these discussions highlight the complex and conserved regulatory network responsible for monitoring the availability and the folding of uS5 for the formation of 40S ribosomal subunits and/or the role of uS5 in potential extra-ribosomal functions.
Keywords: PDCD2; PDCD2L; PRMT3; ZNF277; dedicated chaperone; ribosome biogenesis; uS5.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

