Examining Sporadic Cancer Mutations Uncovers a Set of Genes Involved in Mitochondrial Maintenance
- PMID: 37239369
- PMCID: PMC10218105
- DOI: 10.3390/genes14051009
Examining Sporadic Cancer Mutations Uncovers a Set of Genes Involved in Mitochondrial Maintenance
Abstract
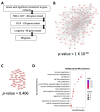
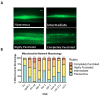
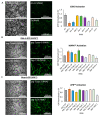
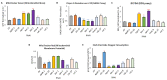
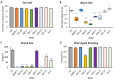
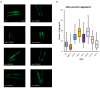
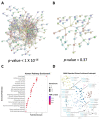
Mitochondria are key organelles for cellular health and metabolism and the activation of programmed cell death processes. Although pathways for regulating and re-establishing mitochondrial homeostasis have been identified over the past twenty years, the consequences of disrupting genes that regulate other cellular processes, such as division and proliferation, on affecting mitochondrial function remain unclear. In this study, we leveraged insights about increased sensitivity to mitochondrial damage in certain cancers, or genes that are frequently mutated in multiple cancer types, to compile a list of candidates for study. RNAi was used to disrupt orthologous genes in the model organism Caenorhabditis elegans, and a series of assays were used to evaluate these genes' importance for mitochondrial health. Iterative screening of ~1000 genes yielded a set of 139 genes predicted to play roles in mitochondrial maintenance or function. Bioinformatic analyses indicated that these genes are statistically interrelated. Functional validation of a sample of genes from this set indicated that disruption of each gene caused at least one phenotype consistent with mitochondrial dysfunction, including increased fragmentation of the mitochondrial network, abnormal steady-state levels of NADH or ROS, or altered oxygen consumption. Interestingly, RNAi-mediated knockdown of these genes often also exacerbated α-synuclein aggregation in a C. elegans model of Parkinson's disease. Additionally, human orthologs of the gene set showed enrichment for roles in human disorders. This gene set provides a foundation for identifying new mechanisms that support mitochondrial and cellular homeostasis.
Keywords: Caenorhabditis elegans; bioinformatics; cancer; development; mitochondria; mitophagy; neurodegeneration; pps-1.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

