Organization and Characterization of the Promoter Elements of the rRNA Operons in the Slow-Growing Pathogen Mycobacterium kumamotonense
- PMID: 37239384
- PMCID: PMC10218544
- DOI: 10.3390/genes14051023
Organization and Characterization of the Promoter Elements of the rRNA Operons in the Slow-Growing Pathogen Mycobacterium kumamotonense
Abstract
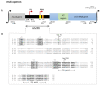
The slow-growing, nontuberculous mycobacterium Mycobacterium kumamotonense possesses two rRNA operons, rrnA and rrnB, located downstream from the murA and tyrS genes, respectively. Here, we report the sequence and organization of the promoter regions of these two rrn operons. In the rrnA operon, transcription can be initiated from the two promoters, named P1 rrnA and PCL1, while in rrnB, transcription can only start from one, called P1 rrnB. Both rrn operons show a similar organization to the one described in Mycobacterium celatum and Mycobacterium smegmatis. Furthermore, by qRT-PCR analyses of the products generated from each promoter, we report that stress conditions such as starvation, hypoxia, and cellular infection affect the contribution of each operon to the synthesis of pre-rRNA. It was found that the products from the PCL1 promoter of rrnA play a pivotal role in rRNA synthesis during all stress conditions. Interestingly, the main participation of the products of transcription from the P1 promoter of rrnB was found during hypoxic conditions at the NRP1 phase. These results provide novel insights into pre-rRNA synthesis in mycobacteria, as well as the potential ability of M. kumamotonense to produce latent infections.
Keywords: Mycobacterium kumamotonense; rRNA operons; rrn promoters.
Conflict of interest statement
The authors declare this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures


References
-
- Masaki T., Ohkusu K., Hata H., Fujiwara N., Iihara H., Yamada-Noda M., Nhung P.H., Hayashi M., Asano Y., Kawamura Y., et al. Mycobacterium kumamotonense Sp. Nov. Recovered from Clinical Specimen and the First Isolation Report of Mycobacterium arupense in Japan: Novel Slowly Growing, Nonchromogenic Clinical Isolates Related to Mycobacterium terrae Complex. Microbiol. Immunol. 2006;50:889–897. doi: 10.1111/j.1348-0421.2006.tb03865.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

