Simultaneous Copy Number Alteration and Single-Nucleotide Variation Analysis in Matched Aqueous Humor and Tumor Samples in Children with Retinoblastoma
- PMID: 37239954
- PMCID: PMC10218537
- DOI: 10.3390/ijms24108606
Simultaneous Copy Number Alteration and Single-Nucleotide Variation Analysis in Matched Aqueous Humor and Tumor Samples in Children with Retinoblastoma
Abstract
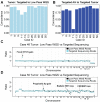
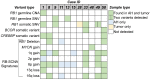
Retinoblastoma (RB) is a childhood cancer that forms in the developing retina of young children; this tumor cannot be biopsied due to the risk of provoking extraocular tumor spread, which dramatically alters the treatment and survival of the patient. Recently, aqueous humor (AH), the clear fluid in the anterior chamber of the eye, has been developed as an organ-specific liquid biopsy for investigation of in vivo tumor-derived information found in the cell-free DNA (cfDNA) of the biofluid. However, identifying somatic genomic alterations, including both somatic copy number alterations (SCNAs) and single nucleotide variations (SNVs) of the RB1 gene, typically requires either: (1) two distinct experimental protocols-low-pass whole genome sequencing for SCNAs and targeted sequencing for SNVs-or (2) expensive deep whole genome or exome sequencing. To save time and cost, we applied a one-step targeted sequencing method to identify both SCNAs and RB1 SNVs in children with RB. High concordance (median = 96.2%) was observed in comparing SCNA calls derived from targeted sequencing to the traditional low-pass whole genome sequencing method. We further applied this method to investigate the degree of concordance of genomic alterations between paired tumor and AH samples from 11 RB eyes. We found 11/11 AH samples (100%) had SCNAs, and 10 of them (90.1%) with recurrent RB-SCNAs, while only nine out of 11 tumor samples (81.8%) had positive RB-SCNA signatures in both low-pass and targeted methods. Eight out of the nine (88.9%) detected SNVs were shared between AH and tumor samples. Ultimately, 11/11 cases have somatic alterations identified, including nine RB1 SNVs and 10 recurrent RB-SCNAs with four focal RB1 deletions and one MYCN gain. The results presented show the feasibility of utilizing one sequencing approach to obtain SCNA and targeted SNV data to capture a broad genomic scope of RB disease, which may ultimately expedite clinical intervention and be less expensive than other methods.
Keywords: RB1 gene; aqueous humor; liquid biopsy; retinoblastoma; somatic copy number alterations; targeted sequencing; variant calling.
Conflict of interest statement
The following authors have no conflict of interest to disclosure: M.J.S., R.K.P., S.P., V.Y., C.P. and P.K.; J.L.B. has financial support from the National Cancer Institute of the National Institute of Health Award Number K08CA232344, The Wright Foundation, Children’s Oncology Group/St. Baldrick’s Foundation, The Knights Templar Eye Foundation, Hyundai Hope on Wheels, Childhood Eye Cancer Trust, Children’s Cancer Research Fund, The Berle & Lucy Adams Chair in Cancer Research, The Larry and Celia Moh Foundation, The Institute for Families, Inc., Children’s Hospital Los Angeles, and an unrestricted departmental grant from Research to Prevent Blindness. L.X has financial support from The Knights Templar Eye Foundation and Children’s Hospital Los Angeles Saban Research Institute, Research Career Development Award J.L.B and L.X. have filed a provisional patent application entitled Aqueous Humor Cell-Free DNA for Diagnostic and Prognostic Evaluation of Ophthalmic Disease 62/654,160 (Berry, Xu, Hicks).
Figures



References
-
- Afshar A.R., Pekmezci M., Bloomer M.M., Cadenas N.J., Stevers M., Banerjee A., Roy R., Olshen A.B., Van Ziffle J., Onodera C., et al. Next-Generation Sequencing of Retinoblastoma Identifies Pathogenic Alterations beyond RB1 Inactivation That Correlate with Aggressive Histopathologic Features. Ophthalmology. 2020;127:804–813. doi: 10.1016/j.ophtha.2019.12.005. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

