NatB Catalytic Subunit Depletion Disrupts DNA Replication Initiation Leading to Senescence in MEFs
- PMID: 37240070
- PMCID: PMC10218504
- DOI: 10.3390/ijms24108724
NatB Catalytic Subunit Depletion Disrupts DNA Replication Initiation Leading to Senescence in MEFs
Abstract
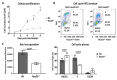
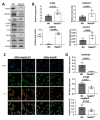
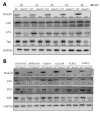
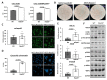
Alpha-aminoterminal acetyltransferase B (NatB) is a critical enzyme responsible for acetylating the aminoterminal end of proteins, thereby modifying approximately 21% of the proteome. This post-translational modification impacts protein folding, structure, stability, and interactions between proteins which, in turn, play a crucial role in modulating several biological functions. NatB has been widely studied for its role in cytoskeleton function and cell cycle regulation in different organisms, from yeast to human tumor cells. In this study, we aimed to understand the biological importance of this modification by inactivating the catalytic subunit of the NatB enzymatic complex, Naa20, in non-transformed mammal cells. Our findings demonstrate that depletion of NAA20 results in decreased cell cycle progression and DNA replication initiation, ultimately leading to the senescence program. Furthermore, we have identified NatB substrates that play a role in cell cycle progression, and their stability is compromised when NatB is inactivated. These results underscore the significance of N-terminal acetylation by NatB in regulating cell cycle progression and DNA replication.
Keywords: DNA replication; N-degron pathway; N-recognins; N-terminal acetylation; NatB; actin cytoskeleton; cell cycle; senescence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Maturation of NAA20 Aminoterminal End Is Essential to Assemble NatB N-Terminal Acetyltransferase Complex.J Mol Biol. 2020 Nov 6;432(22):5889-5901. doi: 10.1016/j.jmb.2020.09.010. Epub 2020 Oct 5. J Mol Biol. 2020. PMID: 32976911
-
NatB-Mediated N-Terminal Acetylation Affects Growth and Biotic Stress Responses.Plant Physiol. 2020 Feb;182(2):792-806. doi: 10.1104/pp.19.00792. Epub 2019 Nov 19. Plant Physiol. 2020. PMID: 31744933 Free PMC article.
-
Physiological importance and identification of novel targets for the N-terminal acetyltransferase NatB.Eukaryot Cell. 2006 Feb;5(2):368-78. doi: 10.1128/EC.5.2.368-378.2006. Eukaryot Cell. 2006. PMID: 16467477 Free PMC article.
-
N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins.J Mol Biol. 2003 Jan 24;325(4):595-622. doi: 10.1016/s0022-2836(02)01269-x. J Mol Biol. 2003. PMID: 12507466 Review.
-
The role of N-acetyltransferases in cancers.Gene. 2024 Jan 20;892:147866. doi: 10.1016/j.gene.2023.147866. Epub 2023 Sep 30. Gene. 2024. PMID: 37783298 Review.
Cited by
-
NatB Protects Procaspase-8 from UBR4-Mediated Degradation and Is Required for Full Induction of the Extrinsic Apoptosis Pathway.Mol Cell Biol. 2024;44(9):358-371. doi: 10.1080/10985549.2024.2382453. Epub 2024 Aug 4. Mol Cell Biol. 2024. PMID: 39099191 Free PMC article.
-
Gene Age Gap Estimate (GAGE) for major depressive disorder: a penalized biological age model using gene expression.bioRxiv [Preprint]. 2024 Nov 17:2024.09.03.610913. doi: 10.1101/2024.09.03.610913. bioRxiv. 2024. Update in: Neurobiol Aging. 2025 Jul;151:13-21. doi: 10.1016/j.neurobiolaging.2025.01.012. PMID: 39282409 Free PMC article. Updated. Preprint.
-
Gene age gap estimate (GAGE) for major depressive disorder: A penalized biological age model using gene expression.Neurobiol Aging. 2025 Jul;151:13-21. doi: 10.1016/j.neurobiolaging.2025.01.012. Epub 2025 Apr 1. Neurobiol Aging. 2025. PMID: 40187167
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

