Synthesis and Degradation of Poly(ADP-ribose) in Zebrafish Brain Exposed to Aluminum
- PMID: 37240112
- PMCID: PMC10218123
- DOI: 10.3390/ijms24108766
Synthesis and Degradation of Poly(ADP-ribose) in Zebrafish Brain Exposed to Aluminum
Abstract
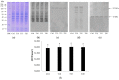
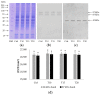
Poly(ADPribosyl)ation is a post-translational protein modification, catalyzed by poly(ADP-ribose) polymerase (PARPs) enzymes, responsible for ADP-ribose polymer synthesis (PAR) from NAD+. PAR turnover is assured by poly(ADPR) glycohydrolase (PARGs) enzymes. In our previous study, the altered histology of zebrafish brain tissue, resulting in demyelination and neurodegeneration also with poly(ADPribosyl)ation hyperactivation, was demonstrated after aluminum (Al) exposure for 10 and 15 days. On the basis of this evidence, the aim of the present research was to study the synthesis and degradation of poly(ADP-ribose) in the brain of adult zebrafish exposed to 11 mg/L of Al for 10, 15, and 20 days. For this reason, PARP and PARG expression analyses were carried out, and ADPR polymers were synthesized and digested. The data showed the presence of different PARP isoforms, among which a human PARP1 counterpart was also expressed. Moreover, the highest PARP and PARG activity levels, responsible for the PAR production and its degradation, respectively, were measured after 10 and 15 days of exposure. We suppose that PARP activation is related to DNA damage induced by Al, while PARG activation is needed to avoid PAR accumulation, which is known to inhibit PARP and promote parthanatos. On the contrary, PARP activity decrease at longer exposure times suggests that neuronal cells could adopt the stratagem of reducing polymer synthesis to avoid energy expenditure and allow cell survival.
Keywords: PARG; PARP; aluminum; brain; neurodegeneration; poly(ADP-ribose); zebrafish.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Poly(ADP-ribose) accumulation and enhancement of postischemic brain damage in 110-kDa poly(ADP-ribose) glycohydrolase null mice.J Cereb Blood Flow Metab. 2006 May;26(5):684-95. doi: 10.1038/sj.jcbfm.9600222. J Cereb Blood Flow Metab. 2006. PMID: 16177811
-
Altered poly(ADP-ribose) metabolism impairs cellular responses to genotoxic stress in a hypomorphic mutant of poly(ADP-ribose) glycohydrolase.Exp Cell Res. 2007 Mar 10;313(5):984-96. doi: 10.1016/j.yexcr.2006.12.025. Epub 2007 Jan 10. Exp Cell Res. 2007. PMID: 17276427
-
Disrupted ADP-ribose metabolism with nuclear Poly (ADP-ribose) accumulation leads to different cell death pathways in presence of hydrogen peroxide in procyclic Trypanosoma brucei.Parasit Vectors. 2016 Mar 23;9:173. doi: 10.1186/s13071-016-1461-1. Parasit Vectors. 2016. PMID: 27007296 Free PMC article.
-
Structure and function of poly(ADP-ribose) polymerase-1: role in oxidative stress-related pathologies.Curr Vasc Pharmacol. 2005 Jul;3(3):209-14. doi: 10.2174/1570161054368625. Curr Vasc Pharmacol. 2005. PMID: 16026317 Review.
-
Poly(ADP-ribose): PARadigms and PARadoxes.Mol Aspects Med. 2013 Dec;34(6):1046-65. doi: 10.1016/j.mam.2012.12.010. Epub 2013 Jan 2. Mol Aspects Med. 2013. PMID: 23290998 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

