A New Opportunity for "Old" Molecules: Targeting PARP1 Activity through a Non-Enzymatic Mechanism
- PMID: 37240195
- PMCID: PMC10219095
- DOI: 10.3390/ijms24108849
A New Opportunity for "Old" Molecules: Targeting PARP1 Activity through a Non-Enzymatic Mechanism
Abstract
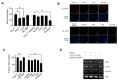
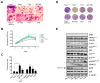
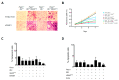
In recent years, new therapies have been developed based on molecules that target molecular mechanisms involved in both the initiation and maintenance of the oncogenic process. Among these molecules are the poly(ADP-ribose) polymerase 1 (PARP1) inhibitors. PARP1 has emerged as a target with great therapeutic potential for some tumor types, drawing attention to this enzyme and resulting in many small molecule inhibitors of its enzymatic activity. Therefore, many PARP inhibitors are currently in clinical trials for the treatment of homologous recombination (HR)-deficient tumors, BRCA-related cancers, taking advantage of synthetic lethality. In addition, several novel cellular functions unrelated to its role in DNA repair have been described, including post-translational modification of transcription factors, or acting through protein-protein interactions as a co-activator or co-repressor of transcription. Previously, we reported that this enzyme may play a key role as a transcriptional co-activator of an important component of cell cycle regulation, the transcription factor E2F1. Here, we show that PARP inhibitors, which interfere with its activity in cell cycle regulation, perform this without affecting its enzymatic function.
Keywords: PARP inhibitors; animal disease models; cancer; neoplasm; poly(ADP-ribose) polymerase-1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Simbulan-Rosenthal C.M., Rosenthal D.S., Luo R., Samara R., Espinoza L.A., Hassa P.O., Hottiger M.O., Smulson M.E. PARP-1 binds E2F-1 independently of its DNA binding and catalytic domains, and acts as a novel coactivator of E2F-1-mediated transcription during re-entry of quiescent cells into S phase. Oncogene. 2003;22:8460–8471. doi: 10.1038/sj.onc.1206897. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

