Electrostatic Screening, Acidic pH and Macromolecular Crowding Increase the Self-Assembly Efficiency of the Minute Virus of Mice Capsid In Vitro
- PMID: 37243141
- PMCID: PMC10224108
- DOI: 10.3390/v15051054
Electrostatic Screening, Acidic pH and Macromolecular Crowding Increase the Self-Assembly Efficiency of the Minute Virus of Mice Capsid In Vitro
Abstract
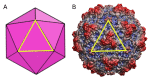
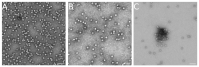
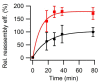
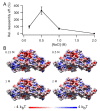
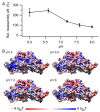
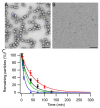
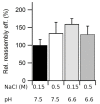
The hollow protein capsids from a number of different viruses are being considered for multiple biomedical or nanotechnological applications. In order to improve the applied potential of a given viral capsid as a nanocarrier or nanocontainer, specific conditions must be found for achieving its faithful and efficient assembly in vitro. The small size, adequate physical properties and specialized biological functions of the capsids of parvoviruses such as the minute virus of mice (MVM) make them excellent choices as nanocarriers and nanocontainers. In this study we analyzed the effects of protein concentration, macromolecular crowding, temperature, pH, ionic strength, or a combination of some of those variables on the fidelity and efficiency of self-assembly of the MVM capsid in vitro. The results revealed that the in vitro reassembly of the MVM capsid is an efficient and faithful process. Under some conditions, up to ~40% of the starting virus capsids were reassembled in vitro as free, non aggregated, correctly assembled particles. These results open up the possibility of encapsidating different compounds in VP2-only capsids of MVM during its reassembly in vitro, and encourage the use of virus-like particles of MVM as nanocontainers.
Keywords: assembly efficiency; capsid; macromolecular crowding; nanocontainer; self-assembly; virus; virus-like particle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

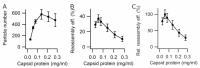

References
-
- Cuervo A., Daudén M.I., Carrascosa J.L. Nucleic acid packaging in viruses. Subcell. Biochem. 2013;68:361–394. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

