Recombinant Virus Quantification Using Single-Cell Droplet Digital PCR: A Method for Infectious Titer Quantification
- PMID: 37243145
- PMCID: PMC10221717
- DOI: 10.3390/v15051060
Recombinant Virus Quantification Using Single-Cell Droplet Digital PCR: A Method for Infectious Titer Quantification
Abstract
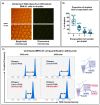
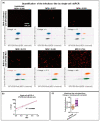
The quantification of viruses is necessary for both research and clinical applications. The methods available for RNA virus quantification possess several drawbacks, including sensitivity to inhibitors and the necessity of a standard curve generation. The main purpose of this study was to develop and validate a method for the quantification of recombinant, replication-deficient Semliki Forest virus (SFV) vectors using droplet digital PCR (ddPCR). This technique demonstrated stability and reproducibility using various sets of primers that targeted inserted transgenes, as well as the nsP1 and nsP4 genes of the SFV genome. Furthermore, the genome titers in the mixture of two types of replication-deficient recombinant virus particles were successfully measured after optimizing the annealing/extension temperature and virus:virus ratios. To measure the infectious units, we developed a single-cell ddPCR, adding the whole infected cells to the droplet PCR mixture. Cell distribution in the droplets was investigated, and β-actin primers were used to normalize the quantification. As a result, the number of infected cells and the virus infectious units were quantified. Potentially, the proposed single-cell ddPCR approach could be used to quantify infected cells for clinical applications.
Keywords: Semliki Forest virus; alphaviruses; ddPCR; infectious titer; replication-deficient virus particles; single-cell ddPCR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Quantitative RT-PCR for titration of replication-defective recombinant Semliki Forest virus.J Virol Methods. 2013 Nov;193(2):647-52. doi: 10.1016/j.jviromet.2013.07.058. Epub 2013 Aug 6. J Virol Methods. 2013. PMID: 23933080
-
Improvement of Precision in Recombinant Adeno-Associated Virus Infectious Titer Assay with Droplet Digital PCR as an Endpoint Measurement.Hum Gene Ther. 2023 Aug;34(15-16):742-757. doi: 10.1089/hum.2023.014. Epub 2023 Jul 4. Hum Gene Ther. 2023. PMID: 37276150 Free PMC article.
-
Optimization of digital droplet polymerase chain reaction for quantification of genetically modified organisms.Biomol Detect Quantif. 2016 Jan 7;7:9-20. doi: 10.1016/j.bdq.2015.12.003. eCollection 2016 Mar. Biomol Detect Quantif. 2016. PMID: 27077048 Free PMC article.
-
Droplet digital PCR as an emerging tool in detecting pathogens nucleic acids in infectious diseases.Clin Chim Acta. 2021 Jun;517:156-161. doi: 10.1016/j.cca.2021.02.008. Epub 2021 Mar 1. Clin Chim Acta. 2021. PMID: 33662358 Review.
-
Droplet digital PCR applications in the tuberculosis world.Tuberculosis (Edinb). 2019 Jul;117:85-92. doi: 10.1016/j.tube.2019.07.001. Epub 2019 Jul 3. Tuberculosis (Edinb). 2019. PMID: 31378274 Review.
Cited by
-
Bacteria and RNA virus inactivation with a high-irradiance UV-A source.Photochem Photobiol Sci. 2024 Oct;23(10):1841-1856. doi: 10.1007/s43630-024-00634-2. Epub 2024 Sep 21. Photochem Photobiol Sci. 2024. PMID: 39305443
-
Macrophage Transcriptomic Alterations Driven by Alphavirus-Based Cancer Immunotherapy Vectors.J Immunol Res. 2025 Jun 13;2025:6573891. doi: 10.1155/jimr/6573891. eCollection 2025. J Immunol Res. 2025. PMID: 40547518 Free PMC article.
-
Digital PCR in Virology: Current Applications and Future Perspectives.Mol Diagn Ther. 2025 Jan;29(1):43-54. doi: 10.1007/s40291-024-00751-9. Epub 2024 Nov 2. Mol Diagn Ther. 2025. PMID: 39487879 Review.
References
-
- Hindson B.J., Ness K.D., Masquelier D.A., Belgrader P., Heredia N.J., Makarewicz A.J., Bright I.J., Lucero M.Y., Hiddessen A.L., Legler T.C., et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011;83:8604. doi: 10.1021/ac202028g. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

