Elucidation of the Cellular Interactome of African Swine Fever Virus Fusion Proteins and Identification of Potential Therapeutic Targets
- PMID: 37243184
- PMCID: PMC10221787
- DOI: 10.3390/v15051098
Elucidation of the Cellular Interactome of African Swine Fever Virus Fusion Proteins and Identification of Potential Therapeutic Targets
Abstract
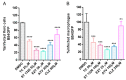
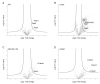
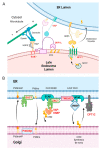
African swine fever virus (ASFV) encodes more than 150 proteins, most of them of unknown function. We used a high-throughput proteomic analysis to elucidate the interactome of four ASFV proteins, which potentially mediate a critical step of the infection cycle, the fusion and endosomal exit of the virions. Using affinity purification and mass spectrometry, we were able to identify potential interacting partners for those ASFV proteins P34, E199L, MGF360-15R and E248R. Representative molecular pathways for these proteins were intracellular and Golgi vesicle transport, endoplasmic reticulum organization, lipid biosynthesis, and cholesterol metabolism. Rab geranyl geranylation emerged as a significant hit, and also Rab proteins, which are crucial regulators of the endocytic pathway and interactors of both p34 and E199L. Rab proteins co-ordinate a tight regulation of the endocytic pathway that is necessary for ASFV infection. Moreover, several interactors were proteins involved in the molecular exchange at ER membrane contacts. These ASFV fusion proteins shared interacting partners, suggesting potential common functions. Membrane trafficking and lipid metabolism were important categories, as we found significant interactions with several enzymes of the lipid metabolism. These targets were confirmed using specific inhibitors with antiviral effect in cell lines and macrophages.
Keywords: ASFV; African swine fever virus; drug target; fusion proteins; interactome; lipid metabolism enzymes; virus–host interaction.
Conflict of interest statement
The authors declare that they have not known competing financial interest or personal relationship that could have appeared to influence the work reported in this paper.
Figures





References
-
- Alonso C., Miskin J., Hernaez B., Fernandez-Zapatero P., Soto L., Canto C., Rodriguez-Crespo I., Dixon L., Escribano J.M. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 2001;75:9819–9827. doi: 10.1128/JVI.75.20.9819-9827.2001. - DOI - PMC - PubMed
-
- Cuesta-Geijo M.A., Garcia-Dorival I., Del Puerto A., Urquiza J., Galindo I., Barrado-Gil L., Lasala F., Cayuela A., Sorzano C.O.S., Gil C., et al. New insights into the role of endosomal proteins for African swine fever virus infection. PLoS Pathog. 2022;18:e1009784. doi: 10.1371/journal.ppat.1009784. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

