In Vitro Antiviral Activity of Nordihydroguaiaretic Acid against SARS-CoV-2
- PMID: 37243241
- PMCID: PMC10222355
- DOI: 10.3390/v15051155
In Vitro Antiviral Activity of Nordihydroguaiaretic Acid against SARS-CoV-2
Abstract
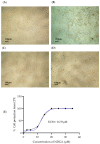
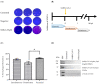
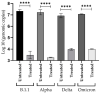
The coronavirus infectious disease 2019 (COVID-19) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and has been spreading rapidly worldwide, creating a pandemic. This article describes the evaluation of the antiviral activity of nordihydroguaiaretic acid (NDGA), a molecule found in Creosote bush (Larrea tridentata) leaves, against SARS-CoV-2 in vitro. A 35 µM concentration of NDGA was not toxic to Vero cells and exhibited a remarkable inhibitory effect on the SARS-CoV-2 cytopathic effect, viral plaque formation, RNA replication, and expression of the SARS-CoV-2 spike glycoprotein. The 50% effective concentration for NDGA was as low as 16.97 µM. Our results show that NDGA could be a promising therapeutic candidate against SARS-CoV-2.
Keywords: Larrea tridentata; NDGA; SARS-CoV-2 antivirals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Virucidal and antiviral activity of astodrimer sodium against SARS-CoV-2 in vitro.Antiviral Res. 2021 Jul;191:105089. doi: 10.1016/j.antiviral.2021.105089. Epub 2021 May 16. Antiviral Res. 2021. PMID: 34010661 Free PMC article.
-
JIB-04 Has Broad-Spectrum Antiviral Activity and Inhibits SARS-CoV-2 Replication and Coronavirus Pathogenesis.mBio. 2022 Feb 22;13(1):e0337721. doi: 10.1128/mbio.03377-21. Epub 2022 Jan 18. mBio. 2022. PMID: 35038906 Free PMC article.
-
Cysteamine with In Vitro Antiviral Activity and Immunomodulatory Effects Has the Potential to Be a Repurposing Drug Candidate for COVID-19 Therapy.Cells. 2021 Dec 24;11(1):52. doi: 10.3390/cells11010052. Cells. 2021. PMID: 35011614 Free PMC article.
-
Molecular mechanisms and clinical applications of nordihydroguaiaretic acid (NDGA) and its derivatives: an update.Med Sci Monit. 2010 May;16(5):RA93-100. Med Sci Monit. 2010. PMID: 20424564 Free PMC article. Review.
-
Nordihydroguaiaretic Acid in Therapeutics: Beneficial to Toxicity Profiles and the Search for its Analogs.Curr Cancer Drug Targets. 2020;20(2):86-103. doi: 10.2174/1568009619666191022141547. Curr Cancer Drug Targets. 2020. PMID: 31642411 Review.
Cited by
-
Nordihydroguaiaretic acid inhibits bladder cancer metastasis through suppression of α1,3-mannosyltransferase expression and LRFN4 N-glycosylation.J Transl Med. 2025 Jul 2;23(1):733. doi: 10.1186/s12967-025-06571-7. J Transl Med. 2025. PMID: 40605099 Free PMC article.
References
-
- Malik Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020;42:3–11. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

