Molecular Epidemiology of SARS-CoV-2 during Five COVID-19 Waves and the Significance of Low-Frequency Lineages
- PMID: 37243279
- PMCID: PMC10223853
- DOI: 10.3390/v15051194
Molecular Epidemiology of SARS-CoV-2 during Five COVID-19 Waves and the Significance of Low-Frequency Lineages
Erratum in
-
Correction: Subramoney et al. Molecular Epidemiology of SARS-CoV-2 during Five COVID-19 Waves and the Significance of Low-Frequency Lineages. Viruses 2023, 15, 1194.Viruses. 2023 Jul 4;15(7):1502. doi: 10.3390/v15071502. Viruses. 2023. PMID: 37515306 Free PMC article.
Abstract
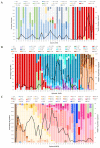
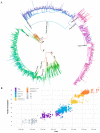
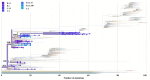
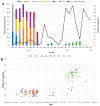
SARS-CoV-2 lineages and variants of concern (VOC) have gained more efficient transmission and immune evasion properties with time. We describe the circulation of VOCs in South Africa and the potential role of low-frequency lineages on the emergence of future lineages. Whole genome sequencing was performed on SARS-CoV-2 samples from South Africa. Sequences were analysed with Nextstrain pangolin tools and Stanford University Coronavirus Antiviral & Resistance Database. In 2020, 24 lineages were detected, with B.1 (3%; 8/278), B.1.1 (16%; 45/278), B.1.1.348 (3%; 8/278), B.1.1.52 (5%; 13/278), C.1 (13%; 37/278) and C.2 (2%; 6/278) circulating during the first wave. Beta emerged late in 2020, dominating the second wave of infection. B.1 and B.1.1 continued to circulate at low frequencies in 2021 and B.1.1 re-emerged in 2022. Beta was outcompeted by Delta in 2021, which was thereafter outcompeted by Omicron sub-lineages during the 4th and 5th waves in 2022. Several significant mutations identified in VOCs were also detected in low-frequency lineages, including S68F (E protein); I82T (M protein); P13L, R203K and G204R/K (N protein); R126S (ORF3a); P323L (RdRp); and N501Y, E484K, D614G, H655Y and N679K (S protein). Low-frequency variants, together with VOCs circulating, may lead to convergence and the emergence of future lineages that may increase transmissibility, infectivity and escape vaccine-induced or natural host immunity.
Keywords: SARS-CoV-2; lineages; low frequency; molecular epidemiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Dhar M.S., Marwal R., Radhakrishnan V.S., Ponnusamy K., Jolly B., Bhoyar R.C., Sardana V., Naushin S., Rophina M., Mellan T.A., et al. Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. Science. 2021;374:995–999. doi: 10.1126/science.abj9932. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

