The TOPLESS corepressor regulates developmental switches in the bryophyte Physcomitrium patens that were critical for plant terrestrialisation
- PMID: 37243383
- PMCID: PMC10953049
- DOI: 10.1111/tpj.16322
The TOPLESS corepressor regulates developmental switches in the bryophyte Physcomitrium patens that were critical for plant terrestrialisation
Abstract
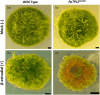
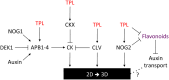
The plant-specific TOPLESS (TPL) family of transcriptional corepressors is integral to multiple angiosperm developmental processes. Despite this, we know little about TPL function in other plants. To address this gap, we investigated the roles TPL plays in the bryophyte Physcomitrium patens, which diverged from angiosperms approximately 0.5 billion years ago. Although complete loss of PpTPL function is lethal, transgenic lines with reduced PpTPL activity revealed that PpTPLs are essential for two fundamental developmental switches in this plant: the transitions from basal photosynthetic filaments (chloronemata) to specialised foraging filaments (caulonemata) and from two-dimensional (2D) to three-dimensional (3D) growth. Using a transcriptomics approach, we integrated PpTPL into the regulatory network governing 3D growth and we propose that PpTPLs represent another important class of regulators that are essential for the 2D-to-3D developmental switch. Transcriptomics also revealed a previously unknown role for PpTPL in the regulation of flavonoids. Intriguingly, 3D growth and the formation of caulonemata were crucial innovations that facilitated the colonisation of land by plants, a major transformative event in the history of life on Earth. We conclude that TPL, which existed before the land plants, was co-opted into new developmental pathways, enabling phytoterrestrialisation and the evolution of land plants.
Keywords: Physcomitrium patens; TOPLESS; land plants; three-dimensional growth; transcriptional corepressor.
© 2023 The Authors. The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Figures




References
-
- Ashton, N.W. & Cove, D.J. (1977) The isolation and preliminary characterization of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Molecular & General Genetics, 154, 87–95.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

