EWSR1 maintains centromere identity
- PMID: 37243594
- PMCID: PMC10758295
- DOI: 10.1016/j.celrep.2023.112568
EWSR1 maintains centromere identity
Abstract
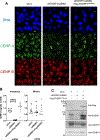
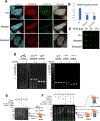
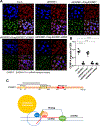
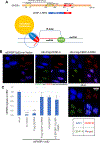
The centromere is essential for ensuring high-fidelity transmission of chromosomes. CENP-A, the centromeric histone H3 variant, is thought to be the epigenetic mark of centromere identity. CENP-A deposition at the centromere is crucial for proper centromere function and inheritance. Despite its importance, the precise mechanism responsible for maintenance of centromere position remains obscure. Here, we report a mechanism to maintain centromere identity. We demonstrate that CENP-A interacts with EWSR1 (Ewing sarcoma breakpoint region 1) and EWSR1-FLI1 (the oncogenic fusion protein in Ewing sarcoma). EWSR1 is required for maintaining CENP-A at the centromere in interphase cells. EWSR1 and EWSR1-FLI1 bind CENP-A through the SYGQ2 region within the prion-like domain, important for phase separation. EWSR1 binds to R-loops through its RNA-recognition motif in vitro. Both the domain and motif are required for maintaining CENP-A at the centromere. Therefore, we conclude that EWSR1 guards CENP-A in centromeric chromatins by binding to centromeric RNA.
Keywords: CENP-A; CENP-A maintenance; CP: Molecular biology; EWSR1; EWSR1-FLI1; Ewing sarcoma; Ewing sarcoma breakpoint region 1; Ewing sarcoma oncogenic fusion protein; centromere; centromere identity; kinetochore; phase separation.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.K. is a guest professor at Osaka University.
Figures



References
-
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, and Bos JL (1988). Genetic alterations during colorectal-tumor development. N Engl J Med 319, 525–532. - PubMed
-
- Hassold TJ, and Jacobs PA (1984). Trisomy in man. Annu Rev Genet 18, 69–97. - PubMed
-
- Kitagawa K, and Hieter P (2001). Evolutionary conservation between budding yeast and human kinetochores. Nat Rev Mol Cell Biol 2, 678–687. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

