IGF2BP1 induces neuroblastoma via a druggable feedforward loop with MYCN promoting 17q oncogene expression
- PMID: 37246217
- PMCID: PMC10226260
- DOI: 10.1186/s12943-023-01792-0
IGF2BP1 induces neuroblastoma via a druggable feedforward loop with MYCN promoting 17q oncogene expression
Abstract
Background: Neuroblastoma is the most common solid tumor in infants accounting for approximately 15% of all cancer-related deaths. Over 50% of high-risk neuroblastoma relapse, emphasizing the need of novel drug targets and therapeutic strategies. In neuroblastoma, chromosomal gains at chromosome 17q, including IGF2BP1, and MYCN amplification at chromosome 2p are associated with adverse outcome. Recent, pre-clinical evidence indicates the feasibility of direct and indirect targeting of IGF2BP1 and MYCN in cancer treatment.
Methods: Candidate oncogenes on 17q were identified by profiling the transcriptomic/genomic landscape of 100 human neuroblastoma samples and public gene essentiality data. Molecular mechanisms and gene expression profiles underlying the oncogenic and therapeutic target potential of the 17q oncogene IGF2BP1 and its cross-talk with MYCN were characterized and validated in human neuroblastoma cells, xenografts and PDX as well as novel IGF2BP1/MYCN transgene mouse models.
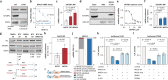
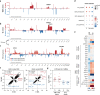
Results: We reveal a novel, druggable feedforward loop of IGF2BP1 (17q) and MYCN (2p) in high-risk neuroblastoma. This promotes 2p/17q chromosomal gains and unleashes an oncogene storm resulting in fostered expression of 17q oncogenes like BIRC5 (survivin). Conditional, sympatho-adrenal transgene expression of IGF2BP1 induces neuroblastoma at a 100% incidence. IGF2BP1-driven malignancies are reminiscent to human high-risk neuroblastoma, including 2p/17q-syntenic chromosomal gains and upregulation of Mycn, Birc5, as well as key neuroblastoma circuit factors like Phox2b. Co-expression of IGF2BP1/MYCN reduces disease latency and survival probability by fostering oncogene expression. Combined inhibition of IGF2BP1 by BTYNB, MYCN by BRD inhibitors or BIRC5 by YM-155 is beneficial in vitro and, for BTYNB, also.
Conclusion: We reveal a novel, druggable neuroblastoma oncogene circuit settling on strong, transcriptional/post-transcriptional synergy of MYCN and IGF2BP1. MYCN/IGF2BP1 feedforward regulation promotes an oncogene storm harboring high therapeutic potential for combined, targeted inhibition of IGF2BP1, MYCN expression and MYCN/IGF2BP1-effectors like BIRC5.
Keywords: 17q gain; BIRC5 (survivin); BTYNB; IGF2BP1; MYCN; Mivebresib; Neuroblastoma; PDX; RNA-binding protein (RBP); Transgenic mouse model; YM-155.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Kushner BH. Neuroblastoma: a disease requiring a multitude of imaging studies. J Nucl Med. 2004;45(7):1172–1188. - PubMed
-
- Bown N, Cotterill S, Lastowska M, O’Neill S, Pearson AD, Plantaz D, Meddeb M, Danglot G, Brinkschmidt C, Christiansen H, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340(25):1954–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

