On the accuracy of the epigenetic copy machine: comprehensive specificity analysis of the DNMT1 DNA methyltransferase
- PMID: 37246710
- PMCID: PMC10359454
- DOI: 10.1093/nar/gkad465
On the accuracy of the epigenetic copy machine: comprehensive specificity analysis of the DNMT1 DNA methyltransferase
Abstract
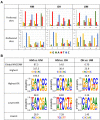
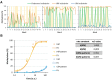
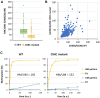
The specificity of DNMT1 for hemimethylated DNA is a central feature for the inheritance of DNA methylation. We investigated this property in competitive methylation kinetics using hemimethylated (HM), hemihydroxymethylated (OH) and unmethylated (UM) substrates with single CpG sites in a randomized sequence context. DNMT1 shows a strong flanking sequence dependent HM/UM specificity of 80-fold on average, which is slightly enhanced on long hemimethylated DNA substrates. To explain this strong effect of a single methyl group, we propose a novel model in which the presence of the 5mC methyl group changes the conformation of the DNMT1-DNA complex into an active conformation by steric repulsion. The HM/OH preference is flanking sequence dependent and on average only 13-fold, indicating that passive DNA demethylation by 5hmC generation is not efficient in many flanking contexts. The CXXC domain of DNMT1 has a moderate flanking sequence dependent contribution to HM/UM specificity during DNA association to DNMT1, but not if DNMT1 methylates long DNA molecules in processive methylation mode. Comparison of genomic methylation patterns from mouse ES cell lines with various deletions of DNMTs and TETs with our data revealed that the UM specificity profile is most related to cellular methylation patterns, indicating that de novo methylation activity of DNMT1 shapes the DNA methylome in these cells.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Allis C.D., Jenuwein T.. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016; 17:487–500. - PubMed
-
- Jeltsch A., Jurkowska R.Z.. New concepts in DNA methylation. Trends Biochem. Sci. 2014; 39:310–318. - PubMed
-
- Schubeler D. Function and information content of DNA methylation. Nature. 2015; 517:321–326. - PubMed
-
- Jeltsch A. Molecular biology. Phylogeny of methylomes. Science. 2010; 328:837–838. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

