Ex vivo to in vivo model of malignant peripheral nerve sheath tumors for precision oncology
- PMID: 37246765
- PMCID: PMC10628938
- DOI: 10.1093/neuonc/noad097
Ex vivo to in vivo model of malignant peripheral nerve sheath tumors for precision oncology
Abstract
Background: Malignant peripheral nerve sheath tumors (MPNST) are aggressive soft tissue sarcomas that often develop in patients with neurofibromatosis type 1 (NF1). To address the critical need for novel therapeutics in MPNST, we aimed to establish an ex vivo 3D platform that accurately captured the genomic diversity of MPNST and could be utilized in a medium-throughput manner for drug screening studies to be validated in vivo using patient-derived xenografts (PDX).
Methods: Genomic analysis was performed on all PDX-tumor pairs. Selected PDX were harvested for assembly into 3D microtissues. Based on prior work in our labs, we evaluated drugs (trabectedin, olaparib, and mirdametinib) ex vivo and in vivo. For 3D microtissue studies, cell viability was the endpoint as assessed by Zeiss Axio Observer. For PDX drug studies, tumor volume was measured twice weekly. Bulk RNA sequencing was performed to identify pathways enriched in cells.
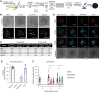
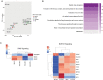
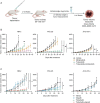
Results: We developed 13 NF1-associated MPNST-PDX and identified mutations or structural abnormalities in NF1 (100%), SUZ12 (85%), EED (15%), TP53 (15%), CDKN2A (85%), and chromosome 8 gain (77%). We successfully assembled PDX into 3D microtissues, categorized as robust (>90% viability at 48 h), good (>50%), or unusable (<50%). We evaluated drug response to "robust" or "good" microtissues, namely MN-2, JH-2-002, JH-2-079-c, and WU-225. Drug response ex vivo predicted drug response in vivo, and enhanced drug effects were observed in select models.
Conclusions: These data support the successful establishment of a novel 3D platform for drug discovery and MPNST biology exploration in a system representative of the human condition.
Keywords: 3D microtissues; MPNST; NF1; PDX; drug screening; genomic variants.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Figures


References
-
- Kourea HP, Bilsky MH, Leung DHY, Lewis JJ, Woodruff JM.. Subdiaphragmatic and intrathoracic paraspinal malignant peripheral nerve sheath tumors: A clinicopathologic study of 25 patients and 26 tumors. Cancer. 1998;82(11):2191–2203. - PubMed
-
- Zheng H, Chang L, Patel N, et al. . Induction of abnormal proliferation by nonmyelinating Schwann cells triggers neurofibroma formation. Cancer Cell. 2008;13(2):117–128. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

