Accelerated Evolution Analysis Uncovers PKNOX2 as a Key Transcription Factor in the Mammalian Cochlea
- PMID: 37247388
- PMCID: PMC10337857
- DOI: 10.1093/molbev/msad128
Accelerated Evolution Analysis Uncovers PKNOX2 as a Key Transcription Factor in the Mammalian Cochlea
Abstract
The genetic bases underlying the evolution of morphological and functional innovations of the mammalian inner ear are poorly understood. Gene regulatory regions are thought to play an important role in the evolution of form and function. To uncover crucial hearing genes whose regulatory machinery evolved specifically in mammalian lineages, we mapped accelerated noncoding elements (ANCEs) in inner ear transcription factor (TF) genes and found that PKNOX2 harbors the largest number of ANCEs within its transcriptional unit. Using reporter gene expression assays in transgenic zebrafish, we determined that four PKNOX2-ANCEs drive differential expression patterns when compared with ortholog sequences from close outgroup species. Because the functional role of PKNOX2 in cochlear hair cells has not been previously investigated, we decided to study Pknox2 null mice generated by CRISPR/Cas9 technology. We found that Pknox2-/- mice exhibit reduced distortion product otoacoustic emissions (DPOAEs) and auditory brainstem response (ABR) thresholds at high frequencies together with an increase in peak 1 amplitude, consistent with a higher number of inner hair cells (IHCs)-auditory nerve synapsis observed at the cochlear basal region. A comparative cochlear transcriptomic analysis of Pknox2-/- and Pknox2+/+ mice revealed that key auditory genes are under Pknox2 control. Hence, we report that PKNOX2 plays a critical role in cochlear sensitivity at higher frequencies and that its transcriptional regulation underwent lineage-specific evolution in mammals. Our results provide novel insights about the contribution of PKNOX2 to normal auditory function and to the evolution of high-frequency hearing in mammals.
Keywords: Ceacam16; PKNOX2; Tectb; hearing; high frequency; mammals.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

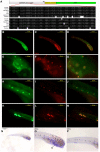
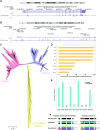
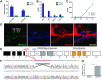
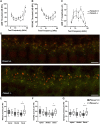

References
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 57: 289–300.
-
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. 1999. Math1: an essential gene for the generation of inner ear hair cells. Science. 284:1837–1841. - PubMed
-
- Bessa J, Tena JJ, de la Calle-Mustienes E, Fernández-Miñán A, Naranjo S, Fernández A, Montoliu L, Akalin A, Lenhard B, Casares F, et al. 2009. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn. 238:2409–2417. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

