Phylogenomic Testing of Root Hypotheses
- PMID: 37247390
- PMCID: PMC10262964
- DOI: 10.1093/gbe/evad096
Phylogenomic Testing of Root Hypotheses
Abstract
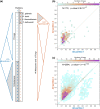
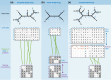
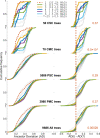
The determination of the last common ancestor (LCA) of a group of species plays a vital role in evolutionary theory. Traditionally, an LCA is inferred by the rooting of a fully resolved species tree. From a theoretical perspective, however, inference of the LCA amounts to the reconstruction of just one branch-the root branch-of the true species tree and should therefore be a much easier task than the full resolution of the species tree. Discarding the reliance on a hypothesized species tree and its rooting leads us to reevaluate what phylogenetic signal is directly relevant to LCA inference and to recast the task as that of sampling the total evidence from all gene families at the genomic scope. Here, we reformulate LCA and root inference in the framework of statistical hypothesis testing and outline an analytical procedure to formally test competing a priori LCA hypotheses and to infer confidence sets for the earliest speciation events in the history of a group of species. Applying our methods to two demonstrative data sets, we show that our inference of the opisthokonta LCA is well in agreement with the common knowledge. Inference of the proteobacteria LCA shows that it is most closely related to modern Epsilonproteobacteria, raising the possibility that it may have been characterized by a chemolithoautotrophic and anaerobic life style. Our inference is based on data comprising between 43% (opisthokonta) and 86% (proteobacteria) of all gene families. Approaching LCA inference within a statistical framework renders the phylogenomic inference powerful and robust.
Keywords: last common ancestor (LCA); phylogenetics; proteobacteria; rooting; species tree.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


References
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 57:289–300.
-
- Campbell BJ, Engel AS, Porter ML, Takai K. 2006. The versatile ε-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol. 4:458–468. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

