From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia
- PMID: 37248212
- PMCID: PMC10227015
- DOI: 10.1038/s41467-023-38668-2
From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia
Abstract
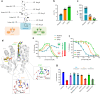
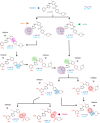
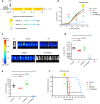
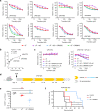
Cancer cells utilize the main de novo pathway and the alternative salvage pathway for deoxyribonucleotide biosynthesis to achieve adequate nucleotide pools. Deoxycytidine kinase is the rate-limiting enzyme of the salvage pathway and it has recently emerged as a target for anti-proliferative therapies for cancers where it is essential. Here, we present the development of a potent inhibitor applying an iterative multidisciplinary approach, which relies on computational design coupled with experimental evaluations. This strategy allows an acceleration of the hit-to-lead process by gradually implementing key chemical modifications to increase affinity and activity. Our lead compound, OR0642, is more than 1000 times more potent than its initial parent compound, masitinib, previously identified from a drug repositioning approach. OR0642 in combination with a physiological inhibitor of the de novo pathway doubled the survival rate in a human T-cell acute lymphoblastic leukemia patient-derived xenograft mouse model, demonstrating the proof-of-concept of this drug design strategy.
© 2023. The Author(s).
Conflict of interest statement
M.S.A., L.H., S.A., K.B.Y., B.S., P.B., E.R., P.R., S.B., P.D., S.C., and X.M. are inventors of a patent entitled “Deoxycytidine kinase inhibitors” filed by the Centre National de la Recherche Scientifique on January 17, 2022 (EP 22 30 5038). The invention relates to the deoxycytidine kinase inhibitors and their uses for treating cancer. The remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

