Rapid evolution of A(H5N1) influenza viruses after intercontinental spread to North America
- PMID: 37248261
- PMCID: PMC10227026
- DOI: 10.1038/s41467-023-38415-7
Rapid evolution of A(H5N1) influenza viruses after intercontinental spread to North America
Abstract
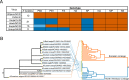
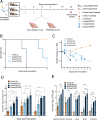
Highly pathogenic avian influenza A(H5N1) viruses of clade 2.3.4.4b underwent an explosive geographic expansion in 2021 among wild birds and domestic poultry across Asia, Europe, and Africa. By the end of 2021, 2.3.4.4b viruses were detected in North America, signifying further intercontinental spread. Here we show that the western movement of clade 2.3.4.4b was quickly followed by reassortment with viruses circulating in wild birds in North America, resulting in the acquisition of different combinations of ribonucleoprotein genes. These reassortant A(H5N1) viruses are genotypically and phenotypically diverse, with many causing severe disease with dramatic neurologic involvement in mammals. The proclivity of the current A(H5N1) 2.3.4.4b virus lineage to reassort and target the central nervous system warrants concerted planning to combat the spread and evolution of the virus within the continent and to mitigate the impact of a potential influenza pandemic that could originate from similar A(H5N1) reassortants.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Smith GJ, Donis RO. World Health Organization/World Organisation for Animal, H. F. & Agriculture Organization, H. E. W. G. Nomenclature updates resulting from the evolution of avian influenza A(H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013-2014. Influenza Other Respir. Viruses. 2015;9:271–276. doi: 10.1111/irv.12324. - DOI - PMC - PubMed
-
- World Health Organization. Antigenic and genetic characteristics of zoonotic influenza A viruses and development of candidate vaccine viruses for pandemic preparedness, https://cdn.who.int/media/docs/default-source/influenza/who-influenza-re... (2022).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

