Natural killer cells and innate lymphoid cells 1 tune anxiety-like behavior and memory in mice via interferon-γ and acetylcholine
- PMID: 37248289
- PMCID: PMC10227025
- DOI: 10.1038/s41467-023-38899-3
Natural killer cells and innate lymphoid cells 1 tune anxiety-like behavior and memory in mice via interferon-γ and acetylcholine
Abstract
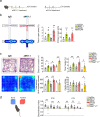
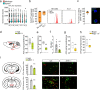
The mechanisms of communication between the brain and the immune cells are still largely unclear. Here, we characterize the populations of resident natural killer (NK) cells and innate lymphoid cells (ILC) 1 in the meningeal dura layer of adult mice. We describe that ILC1/NK cell-derived interferon-γ and acetylcholine can contribute to the modulation of brain homeostatic functions, shaping synaptic neuronal transmission and neurotransmitter levels with effects on mice behavior. In detail, the interferon-γ plays a role in the formation of non-spatial memory, tuning the frequency of GABAergic neurotransmission on cortical pyramidal neurons, while the acetylcholine is a mediator involved in the modulation of brain circuitries that regulate anxiety-like behavior. These findings disclose mechanisms of immune-to-brain communication that modulate brain functions under physiological conditions.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

