Limiting mitochondrial plasticity by targeting DRP1 induces metabolic reprogramming and reduces breast cancer brain metastases
- PMID: 37248394
- PMCID: PMC11290463
- DOI: 10.1038/s43018-023-00563-6
Limiting mitochondrial plasticity by targeting DRP1 induces metabolic reprogramming and reduces breast cancer brain metastases
Abstract
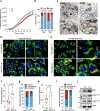
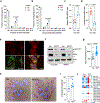
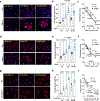
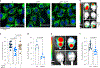
Disseminated tumor cells with metabolic flexibility to utilize available nutrients in distal organs persist, but the precise mechanisms that facilitate metabolic adaptations remain unclear. Here we show fragmented mitochondrial puncta in latent brain metastatic (Lat) cells enable fatty acid oxidation (FAO) to sustain cellular bioenergetics and maintain redox homeostasis. Depleting the enriched dynamin-related protein 1 (DRP1) and limiting mitochondrial plasticity in Lat cells results in increased lipid droplet accumulation, impaired FAO and attenuated metastasis. Likewise, pharmacological inhibition of DRP1 using a small-molecule brain-permeable inhibitor attenuated metastatic burden in preclinical models. In agreement with these findings, increased phospho-DRP1 expression was observed in metachronous brain metastasis compared with patient-matched primary tumors. Overall, our findings reveal the pivotal role of mitochondrial plasticity in supporting the survival of Lat cells and highlight the therapeutic potential of targeting cellular plasticity programs in combination with tumor-specific alterations to prevent metastatic recurrences.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
A.B.H. has received research grant support from Takeda and travel support from Puma Biotechnology. C.L.A. receives or has received research grant support from Pfizer, Lilly and Takeda; holds stock options in Provista; and serves or has served in an advisory role to Novartis, Lilly, TAIHO Oncology, Daiichi Sankyo, Merck, AstraZeneca, OrigiMed, Immunomedics and Susan G. Komen Foundation. R.J.D. is a founder of Atavistik Biosciences and an advisor for Agios Pharmaceuticals, Nirogy Therapeutics and Vida Ventures. The other authors declare no competing interests.
Figures













Comment in
-
Feeding latent brain metastasis.Nat Rev Cancer. 2023 Aug;23(8):510. doi: 10.1038/s41568-023-00601-6. Nat Rev Cancer. 2023. PMID: 37369834 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

