Monocyte-derived IL-6 programs microglia to rebuild damaged brain vasculature
- PMID: 37248420
- PMCID: PMC11531796
- DOI: 10.1038/s41590-023-01521-1
Monocyte-derived IL-6 programs microglia to rebuild damaged brain vasculature
Abstract
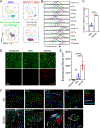
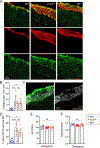
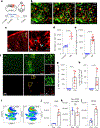
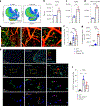
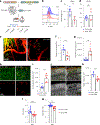
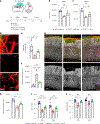
Cerebrovascular injury (CVI) is a common pathology caused by infections, injury, stroke, neurodegeneration and autoimmune disease. Rapid resolution of a CVI requires a coordinated innate immune response. In the present study, we sought mechanistic insights into how central nervous system-infiltrating monocytes program resident microglia to mediate angiogenesis and cerebrovascular repair after an intracerebral hemorrhage. In the penumbrae of human stroke brain lesions, we identified a subpopulation of microglia that express vascular endothelial growth factor A. These cells, termed 'repair-associated microglia' (RAMs), were also observed in a rodent model of CVI and coexpressed interleukin (IL)-6Ra. Cerebrovascular repair did not occur in IL-6 knockouts or in mice lacking microglial IL-6Ra expression and single-cell transcriptomic analyses revealed faulty RAM programming in the absence of IL-6 signaling. Infiltrating CCR2+ monocytes were the primary source of IL-6 after a CVI and were required to endow microglia with proliferative and proangiogenic properties. Faulty RAM programming in the absence of IL-6 or inflammatory monocytes resulted in poor cerebrovascular repair, neuronal destruction and sustained neurological deficits that were all restored via exogenous IL-6 administration. These data provide a molecular and cellular basis for how monocytes instruct microglia to repair damaged brain vasculature and promote functional recovery after injury.
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures








Similar articles
-
CX3CR1-CCR2-dependent monocyte-microglial signaling modulates neurovascular leakage and acute injury in a mouse model of childhood stroke.J Cereb Blood Flow Metab. 2019 Oct;39(10):1919-1935. doi: 10.1177/0271678X18817663. Epub 2019 Jan 10. J Cereb Blood Flow Metab. 2019. PMID: 30628839 Free PMC article.
-
Functional differences between microglia and monocytes after ischemic stroke.J Neuroinflammation. 2015 May 29;12:106. doi: 10.1186/s12974-015-0329-1. J Neuroinflammation. 2015. PMID: 26022493 Free PMC article.
-
IL-33/ST2 signaling in monocyte-derived macrophages maintains blood-brain barrier integrity and restricts infarctions early after ischemic stroke.J Neuroinflammation. 2024 Oct 24;21(1):274. doi: 10.1186/s12974-024-03264-8. J Neuroinflammation. 2024. PMID: 39449077 Free PMC article.
-
Functional consequences of a close encounter between microglia and brain-infiltrating monocytes during CNS pathology and repair.J Leukoc Biol. 2021 Jul;110(1):89-106. doi: 10.1002/JLB.3RU0820-536R. Epub 2020 Nov 6. J Leukoc Biol. 2021. PMID: 33155726 Review.
-
Cellular players that shape evolving pathology and neurodegeneration following traumatic brain injury.Brain Behav Immun. 2018 Jul;71:9-17. doi: 10.1016/j.bbi.2018.03.033. Epub 2018 Mar 27. Brain Behav Immun. 2018. PMID: 29601944 Review.
Cited by
-
Possible Impact of Peripheral Inflammatory Factors and Interleukin-1β (IL-1β) on Cognitive Functioning in Progressive Supranuclear Palsy-Richardson Syndrome (PSP-RS) and Progressive Supranuclear Palsy-Predominant Parkinsonism (PSP-P).Int J Mol Sci. 2024 Dec 9;25(23):13211. doi: 10.3390/ijms252313211. Int J Mol Sci. 2024. PMID: 39684921 Free PMC article.
-
Polymorphisms in genes related to inflammation and endothelial function are associated with ischemic stroke and other vascular events in populations at high risk of stroke.J Cent Nerv Syst Dis. 2025 Jan 3;17:11795735241312660. doi: 10.1177/11795735241312660. eCollection 2025. J Cent Nerv Syst Dis. 2025. PMID: 39763503 Free PMC article.
-
Unlocking the therapeutic potential of tumor-derived EVs in ischemia-reperfusion: a breakthrough perspective from glioma and stroke.J Neuroinflammation. 2025 Mar 15;22(1):84. doi: 10.1186/s12974-025-03405-7. J Neuroinflammation. 2025. PMID: 40089793 Free PMC article. Review.
-
Mechanism of disturbed endothelial cell function on angiogenesis following ischemic brain stroke (Review).Exp Ther Med. 2025 Jan 29;29(4):61. doi: 10.3892/etm.2025.12811. eCollection 2025 Apr. Exp Ther Med. 2025. PMID: 39991719 Free PMC article. Review.
-
NO-releasing double-crosslinked responsive hydrogels accelerate the treatment and repair of ischemic stroke.Acta Pharm Sin B. 2025 Feb;15(2):1112-1125. doi: 10.1016/j.apsb.2025.01.005. Epub 2025 Jan 20. Acta Pharm Sin B. 2025. PMID: 40177574 Free PMC article.
References
-
- Kenney K et al. Cerebral vascular injury in traumatic brain injury. Exp. Neurol. 275, 353–366 (2016). - PubMed
-
- Badjatia N & Rosand J Intracerebral hemorrhage. Neurologist 11, 311–324 (2005). - PubMed
-
- Twitchell TE The restoration of motor function following hemiplegia in man. Brain 74, 443–480 (1951). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

