Vinculin Y822 is an important determinant of ligand binding
- PMID: 37248996
- PMCID: PMC10281268
- DOI: 10.1242/jcs.260104
Vinculin Y822 is an important determinant of ligand binding
Abstract
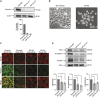
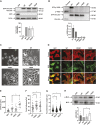
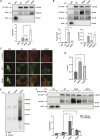
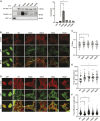
Vinculin is an actin-binding protein present at cell-matrix and cell-cell adhesions, which plays a critical role in bearing force experienced by cells and dissipating it onto the cytoskeleton. Recently, we identified a key tyrosine residue, Y822, whose phosphorylation plays a critical role in force transmission at cell-cell adhesions. The role of Y822 in human cancer remains unknown, even though Y822 is mutated to Y822C in uterine cancers. Here, we investigated the effect of this amino acid substitution and that of a phosphodeficient Y822F vinculin in cancer cells. We observed that the presence of the Y822C mutation led to cells that proliferate and migrate more rapidly and contained smaller focal adhesions when compared to cells with wild-type vinculin. In contrast, the presence of the Y822F mutation led to highly spread cells with larger focal adhesions and increased contractility. Furthermore, we provide evidence that Y822C vinculin forms a disulfide bond with paxillin, accounting for some of the elevated phosphorylated paxillin recruitment. Taken together, these data suggest that vinculin Y822 modulates the recruitment of ligands.
Keywords: Cell–cell adhesion; Cell–matrix adhesion; Mechanotransduction; Vinculin.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

References
-
- Abé, C., Dietrich, F., Gajula, P., Benz, M., Vogel, K.-P., Van Gastel, M., Illenberger, S., Ziegler, W. H. and Steinhoff, H.-J. (2011). Monomeric and dimeric conformation of the vinculin tail five-helix bundle in solution studied by EPR spectroscopy. Biophys. J. 101, 1772-1780. 10.1016/j.bpj.2011.08.048 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

