Whole-Genome Subtyping Reveals Population Structure and Host Adaptation of Salmonella Typhimurium from Wild Birds
- PMID: 37249426
- PMCID: PMC10281135
- DOI: 10.1128/jcm.01847-22
Whole-Genome Subtyping Reveals Population Structure and Host Adaptation of Salmonella Typhimurium from Wild Birds
Abstract
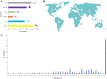
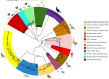
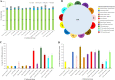
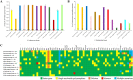
Within-host evolution of bacterial pathogens can lead to host-associated variants of the same species or serovar. Identification and characterization of closely related variants from diverse host species are crucial to public health and host-pathogen adaptation research. However, the work remained largely underexplored at a strain level until the advent of whole-genome sequencing (WGS). Here, we performed WGS-based subtyping and analyses of Salmonella enterica serovar Typhimurium (n = 787) from different wild birds across 18 countries over a 75-year period. We revealed seven avian host-associated S. Typhimurium variants/lineages. These lineages emerged globally over short timescales and presented genetic features distinct from S. Typhimurium lineages circulating among humans and domestic animals. We further showed that, in terms of virulence, host adaptation of these variants was driven by genome degradation. Our results provide a snapshot of the population structure and genetic diversity of S. Typhimurium within avian hosts. We also demonstrate the value of WGS-based subtyping and analyses in unravelling closely related variants at the strain level.
Keywords: Salmonella enterica serovar Typhimurium; avian hosts; host adaptation; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Stanaway JD, Parisi A, Sarkar K, Blacker BF, Reiner RC, Hay SI, Nixon MR, Dolecek C, James SL, Mokdad AH, Abebe G, Ahmadian E, Alahdab F, Alemnew BTT, Alipour V, Allah Bakeshei F, Animut MD, Ansari F, Arabloo J, Asfaw ET, Bagherzadeh M, Bassat Q, Belayneh YMM, Carvalho F, Daryani A, Demeke FM, Demis ABB, Dubey M, Duken EE, Dunachie SJ, Eftekhari A, Fernandes E, Fouladi Fard R, Gedefaw GA, Geta B, Gibney KB, Hasanzadeh A, Hoang CL, Kasaeian A, Khater A, Kidanemariam ZT, Lakew AM, Malekzadeh R, Melese A, Mengistu DT, Mestrovic T, Miazgowski B, Mohammad KA, Mohammadian M, Mohammadian-Hafshejani A, et al. 2019. The global burden of non-typhoidal Salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis 19:1312–1324. doi: 10.1016/S1473-3099(19)30418-9. - DOI - PMC - PubMed
-
- Grimont PA, Weill FX. 2007. Antigenic formulae of the Salmonella serovars, 9th ed. WHO; Collaborating Centre for Salmonella: Paris, France.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

