Peptidomics
- PMID: 37250919
- PMCID: PMC7614574
- DOI: 10.1038/s43586-023-00205-2
Peptidomics
Abstract
Peptides are biopolymers, typically consisting of 2-50 amino acids. They are biologically produced by the cellular ribosomal machinery or by non-ribosomal enzymes and, sometimes, other dedicated ligases. Peptides are arranged as linear chains or cycles, and include post-translational modifications, unusual amino acids and stabilizing motifs. Their structure and molecular size render them a unique chemical space, between small molecules and larger proteins. Peptides have important physiological functions as intrinsic signalling molecules, such as neuropeptides and peptide hormones, for cellular or interspecies communication, as toxins to catch prey or as defence molecules to fend off enemies and microorganisms. Clinically, they are gaining popularity as biomarkers or innovative therapeutics; to date there are more than 60 peptide drugs approved and more than 150 in clinical development. The emerging field of peptidomics comprises the comprehensive qualitative and quantitative analysis of the suite of peptides in a biological sample (endogenously produced, or exogenously administered as drugs). Peptidomics employs techniques of genomics, modern proteomics, state-of-the-art analytical chemistry and innovative computational biology, with a specialized set of tools. The complex biological matrices and often low abundance of analytes typically examined in peptidomics experiments require optimized sample preparation and isolation, including in silico analysis. This Primer covers the combination of techniques and workflows needed for peptide discovery and characterization and provides an overview of various biological and clinical applications of peptidomics.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures


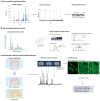
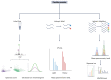

References
-
- Dang T, Süssmuth RD. Bioactive peptide natural products as lead structures for medicinal use. Acc Chem Res. 2017;50:1566–1576. - PubMed
-
- Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov Today. 2015;20:122–128. - PubMed
-
- Muttenthaler M, King GF, Adams DJ, Alewood PF. Trends in peptide drug discovery. Nat Rev Drug Discov. 2021;20:309–325. [This comprehensive review discusses the importance of peptides as drug leads and innovative therapeutics.] - PubMed
-
- Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81:136–147. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
